Longitudinal omics data and preclinical treatment suggest the proteasome inhibitor carfilzomib as therapy for ibrutinib-resistant CLL
- PMID: 39863584
- PMCID: PMC11762753
- DOI: 10.1038/s41467-025-56318-7
Longitudinal omics data and preclinical treatment suggest the proteasome inhibitor carfilzomib as therapy for ibrutinib-resistant CLL
Abstract
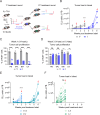
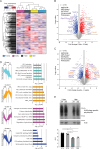
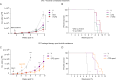
Chronic lymphocytic leukemia is a malignant lymphoproliferative disorder for which primary or acquired drug resistance represents a major challenge. To investigate the underlying molecular mechanisms, we generate a mouse model of ibrutinib resistance, in which, after initial treatment response, relapse under therapy occurrs with an aggressive outgrowth of malignant cells, resembling observations in patients. A comparative analysis of exome, transcriptome and proteome of sorted leukemic murine cells during treatment and after relapse suggests alterations in the proteasome activity as a driver of ibrutinib resistance. Preclinical treatment with the irreversible proteasome inhibitor carfilzomib administered upon ibrutinib resistance prolongs survival of mice. Longitudinal proteomic analysis of ibrutinib-resistant patients identifies deregulation in protein post-translational modifications. Additionally, cells from ibrutinib-resistant patients effectively respond to several proteasome inhibitors in co-culture assays. Altogether, our results from orthogonal omics approaches identify proteasome inhibition as potentially attractive treatment for chronic lymphocytic leukemia patients resistant or refractory to ibrutinib.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: S.S.S. has received honoraria from AbbVie, AstraZeneca, BeiGene, and Janssen, and research support from BeiGene and TG Therapeutics outside of this work. E.T. has received honoraria from Abbvie, Janssen, AstraZeneca, BeiGene and research support by Abbvie, Hofmann-La Roche and Janssen-Cilag. StSt has received advisory board honoraria, research support, travel support, speaker fees from AbbVie, AstraZeneca, BeiGene, BMS, Gilead, GSK, Hoffmann-La Roche, Janssen, Novartis, and Sunesis. JD has received research funding from Roche/Genentech. MS has received advisory board honoraria from AbbVie and research support from Bayer AG. The remaining authors declare no competing interests.
Figures




References
-
- Nass, J. & Efferth, T. Drug targets and resistance mechanisms in multiple myeloma. Cancer Drug Resistance1, 87–117 (2018).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

