Mathematical model linking telomeres to senescence in Saccharomyces cerevisiae reveals cell lineage versus population dynamics
- PMID: 39863614
- PMCID: PMC11762778
- DOI: 10.1038/s41467-025-56196-z
Mathematical model linking telomeres to senescence in Saccharomyces cerevisiae reveals cell lineage versus population dynamics
Abstract
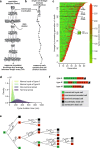
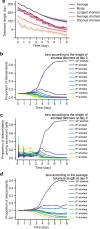
Telomere shortening ultimately causes replicative senescence. However, identifying the mechanisms driving replicative senescence in cell populations is challenging due to the heterogeneity of telomere lengths and the asynchrony of senescence onset. Here, we present a mathematical model of telomere shortening and replicative senescence in Saccharomyces cerevisiae which is quantitatively calibrated and validated using data of telomerase-deficient single cells. Simulations of yeast populations, where cells with varying proliferation capacities compete against each other, show that the distribution of telomere lengths of the initial population shapes population growth, especially through the distribution of cells' shortest telomere lengths. We also quantified how factors influencing cell viability independently of telomeres can impact senescence rates. Overall, we demonstrate a temporal evolution in the composition of senescent cell populations-from a state directly linked to critically short telomeres to a state where senescence onset becomes stochastic. This population structure may promote genome instability and facilitate senescence escape.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






Similar articles
-
The asymmetry of telomere replication contributes to replicative senescence heterogeneity.Sci Rep. 2015 Oct 15;5:15326. doi: 10.1038/srep15326. Sci Rep. 2015. PMID: 26468778 Free PMC article.
-
The length of the shortest telomere as the major determinant of the onset of replicative senescence.Genetics. 2013 Aug;194(4):847-57. doi: 10.1534/genetics.113.152322. Epub 2013 Jun 3. Genetics. 2013. PMID: 23733785 Free PMC article.
-
A subtelomeric region affects telomerase-negative replicative senescence in Saccharomyces cerevisiae.Sci Rep. 2019 Feb 12;9(1):1845. doi: 10.1038/s41598-018-38000-9. Sci Rep. 2019. PMID: 30755624 Free PMC article.
-
The many types of heterogeneity in replicative senescence.Yeast. 2019 Nov;36(11):637-648. doi: 10.1002/yea.3433. Epub 2019 Aug 6. Yeast. 2019. PMID: 31306505 Free PMC article. Review.
-
Telomere length homeostasis.Chromosoma. 2006 Dec;115(6):413-25. doi: 10.1007/s00412-006-0067-3. Epub 2006 Jun 2. Chromosoma. 2006. PMID: 16741708 Review.
References
-
- Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell144, 646–674 (2011). - PubMed
-
- Campisi, J. & d'Adda di Fagagna, F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol.8, 729–740 (2007). - PubMed
-
- Bryan, T. M., Englezou, A., Dalla-Pozza, L., Dunham, M. A. & Reddel, R. R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med3, 1271–1274 (1997). - PubMed
MeSH terms
Substances
Grants and funding
- ANR-16-CE12-0026/Agence Nationale de la Recherche (French National Research Agency)
- ANR-11-LABX-0011-01/Agence Nationale de la Recherche (French National Research Agency)
- PLBIO20-312 INCa_15192/Institut National Du Cancer (French National Cancer Institute)
- Equipe Labellisée/Fondation pour la Recherche Médicale (Foundation for Medical Research in France)
- SKIPPERAD 306321/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
LinkOut - more resources
Full Text Sources

