Mucus-penetrating microbiota drive chronic low-grade intestinal inflammation and metabolic dysregulation
- PMID: 39865067
- PMCID: PMC11776472
- DOI: 10.1080/19490976.2025.2455790
Mucus-penetrating microbiota drive chronic low-grade intestinal inflammation and metabolic dysregulation
Abstract
Metabolic syndrome is, in humans, associated with alterations in the composition and localization of the intestinal microbiota, including encroachment of bacteria within the colon's inner mucus layer. Possible promoters of these events include dietary emulsifiers, such as carboxymethylcellulose (CMC) and polysorbate-80 (P80), which, in mice, result in altered microbiota composition, encroachment, low-grade inflammation and metabolic syndrome. While assessments of gut microbiota composition have largely focused on fecal/luminal samples, we hypothesize an outsized role for changes in mucus microbiota in driving low-grade inflammation and its consequences. In support of this notion, we herein report that both CMC and P80 led to stark changes in the mucus microbiome, markedly distinct from those observed in feces. Moreover, transfer of mucus microbiota from CMC- and P80-fed mice to germfree mice resulted in microbiota encroachment, low-grade inflammation, and various features of metabolic syndrome. Thus, we conclude that mucus-associated bacteria are pivotal determinants of intestinal inflammatory tone and host metabolism.
Keywords: Microbiota; encroachment; inflammation; metabolic deregulations; mucus.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
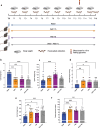
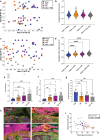

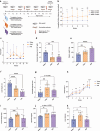
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
