The cryptic lncRNA-encoded microprotein TPM3P9 drives oncogenic RNA splicing and tumorigenesis
- PMID: 39865075
- PMCID: PMC11770092
- DOI: 10.1038/s41392-025-02128-8
The cryptic lncRNA-encoded microprotein TPM3P9 drives oncogenic RNA splicing and tumorigenesis
Abstract
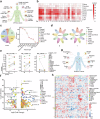
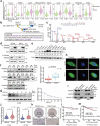
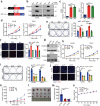
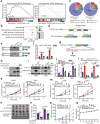
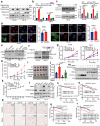
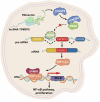
Emerging evidence demonstrates that cryptic translation from RNAs previously annotated as noncoding might generate microproteins with oncogenic functions. However, the importance and underlying mechanisms of these microproteins in alternative splicing-driven tumor progression have rarely been studied. Here, we show that the novel protein TPM3P9, encoded by the lncRNA tropomyosin 3 pseudogene 9, exhibits oncogenic activity in clear cell renal cell carcinoma (ccRCC) by enhancing oncogenic RNA splicing. Overexpression of TPM3P9 promotes cell proliferation and tumor growth. Mechanistically, TPM3P9 binds to the RRM1 domain of the splicing factor RBM4 to inhibit RBM4-mediated exon skipping in the transcription factor TCF7L2. This results in increased expression of the oncogenic splice variant TCF7L2-L, which activates NF-κB signaling via its interaction with SAM68 to transcriptionally induce RELB expression. From a clinical perspective, TPM3P9 expression is upregulated in cancer tissues and is significantly correlated with the expression of TCF7L2-L and RELB. High TPM3P9 expression or low RBM4 expression is associated with poor survival in patients with ccRCC. Collectively, our findings functionally and clinically characterize the "noncoding RNA"-derived microprotein TPM3P9 and thus identify potential prognostic and therapeutic factors in renal cancer.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Venter, J. C. et al. The sequence of the human genome. Science291, 1304–1351 (2001). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

