Composition and liquid-to-solid maturation of protein aggregates contribute to bacterial dormancy development and recovery
- PMID: 39865082
- PMCID: PMC11770139
- DOI: 10.1038/s41467-025-56387-8
Composition and liquid-to-solid maturation of protein aggregates contribute to bacterial dormancy development and recovery
Abstract
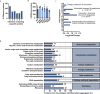
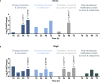
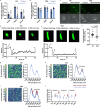
Recalcitrant bacterial infections can be caused by various types of dormant bacteria, including persisters and viable but nonculturable (VBNC) cells. Despite their clinical importance, we know fairly little about bacterial dormancy development and recovery. Previously, we established a correlation between protein aggregation and dormancy in Escherichia coli. Here, we present further support for a direct relationship between both. Our experiments demonstrate that aggregates progressively sequester proteins involved in energy production, thereby likely causing ATP depletion and dormancy. Furthermore, we demonstrate that structural features of protein aggregates determine the cell's ability to exit dormancy and resume growth. Proteins were shown to first assemble in liquid-like condensates that solidify over time. This liquid-to-solid phase transition impedes aggregate dissolution, thereby preventing growth resumption. Our data support a model in which aggregate structure, rather than cellular activity, marks the transition from the persister to the VBNC state.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Darby, E. M. et al. Molecular mechanisms of antibiotic resistance revisited. Nat Rev Microbiol21, 280–295 (2023). - PubMed
-
- Fauvart, M., de Groote, V. N. & Michiels, J. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol60, 699–709 (2011). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

