Overexpression of miR-124 enhances the therapeutic benefit of TMZ treatment in the orthotopic GBM mice model by inhibition of DNA damage repair
- PMID: 39865088
- PMCID: PMC11770086
- DOI: 10.1038/s41419-025-07363-z
Overexpression of miR-124 enhances the therapeutic benefit of TMZ treatment in the orthotopic GBM mice model by inhibition of DNA damage repair
Abstract
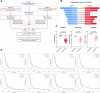
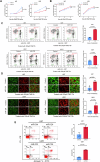
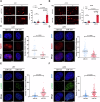
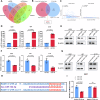
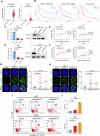
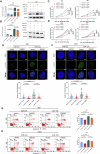
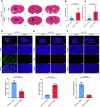
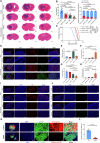
Glioblastoma (GBM) is the most common malignant primary brain cancer with poor prognosis due to the resistant to current treatments, including the first-line drug temozolomide (TMZ). Accordingly, it is urgent to clarify the mechanism of chemotherapeutic resistance to improve the survival rate of patients. In the present study, by integrating comprehensive non-coding RNA-seq data from multiple cohorts of GBM patients, we identified that a series of miRNAs are frequently downregulated in GBM patients compared with the control samples. Among them, a high level of miR-124 is closely associated with a favorable survival rate in the clinical patients. In the phenotype experiment, we demonstrated that miR-124 overexpression increases responsiveness of GBM cells to TMZ-induced cell death, and vice versa. In the mechanistic study, we for the first time identified that RAD51, a key functional molecule in DNA damage repair, is a novel and bona fide target of miR-124 in GBM cells. Given that other miR-124-regulated mechanisms on TMZ sensitivity have been reported, we performed recue experiment to demonstrate that RAD51 is essential for miR-124-mediated sensitivity to TMZ in GBM cells. More importantly, our in vivo functional experiment showed that combinational utilization of miR-124 overexpression and TMZ presents a synergetic therapeutic benefit in the orthotopic GBM mice model. Taken together, we rationally explained a novel and important mechanism of the miR-124-mediated high sensitivity to TMZ-induced cell death in GBM and provided evidence to support that miR-124-RAD51 regulatory axis could be a promising candidate in the comprehensive treatment with TMZ in GBM.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethical approval: The protocol for the animal study was approved by the Animal Welfare and Ethics Committee of the Fourth Military Medical University (Xi’an, China).
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

