Comprehensive analysis reveals cholesterol metabolism-related signature for predicting prognosis and guiding individualized treatment of glioma
- PMID: 39866460
- PMCID: PMC11757779
- DOI: 10.1016/j.heliyon.2024.e41601
Comprehensive analysis reveals cholesterol metabolism-related signature for predicting prognosis and guiding individualized treatment of glioma
Abstract
Objective: Gliomas are the most common intracranial tumors with the highest degree of malignancy. Disturbed cholesterol metabolism is one of the key features of many malignant tumors, including gliomas. This study aimed to investigate the significance of cholesterol metabolism-related genes in prognostic prediction and in guiding individualized treatment of patients with gliomas.
Methods: Transcriptional data and clinicopathological data were obtained from The Cancer Genome Atlas (TCGA) and Chinese Glioma Genome Atlas (CGGA) databases. Intraoperative glioma samples retained in our unit and the corresponding clinicopathological information were also collected with the patients' knowledge. Firstly, cholesterol metabolism-related gene signatures (CMRGS) were identified and constructed based on difference analysis, least absolute shrinkage and selection operator (LASSO) regression analysis, and univariate/multivariate COX analysis. Then, the role of CMRGS in predicting the prognosis of gliomas and distinguishing immune landscapes was evaluated by using nomograms, survival analysis, enrichment analysis, and immune-infiltration analysis. Finally, the drug sensitivity of gliomas in different risk groups was evaluated using the oncoPredict algorithm, and potentially sensitive chemotherapeutic and molecular-targeted drugs were identified.
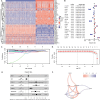
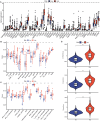
Results: The prognostic CMRGS contained seven genes: APOE, SCD, CXCL16, FABP5, S100A11, TNFRSF12A, and ELOVL2. Patients were divided into high- and low-risk groups based on the median cholesterol metabolic index (CMI). There were significant differences in clinicopathological characteristics and overall survival between groups. COX analysis suggested that CMRGS was an independent risk factor for glioma prognosis and had a better predictive performance than several classical indicators. In addition, GSEA, immune infiltration analysis showed that CMRGS could differentiate the immune landscapes of patients in groups. The reliability of CMRGS was validated in the CGGA cohort and our Gusu cohort. Finally, 14 drugs sensitive to high-risk patients and 16 drugs sensitive to low-risk patients were identified.
Conclusion: The CMRGS reliably predicts glioma prognosis in multiple cohorts and may be useful in guiding individualized treatment.
Keywords: Cholesterol metabolism; Drug sensitivity; Glioma; Immunoscape; Prognostic prediction; Signature.
© 2024 Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






References
-
- Xu S., Tang L., Li X., Fan F., Liu Z. Immunotherapy for glioma: current management and future application. Cancer Lett. 2020;476 - PubMed
-
- Weller M., Le Rhun E. How did lomustine become standard of care in recurrent glioblastoma? Cancer Treat Rev. 2020;87 - PubMed
-
- Cheng M., Zhang Z.W., Ji X.H., Xu Y., Bian E., Zhao B. Super-enhancers: a new frontier for glioma treatment. Biochim. Biophys. Acta Rev. Canc. 2020;1873(2) - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

