UVREK: Development and Analysis of an Expression Profile Knowledgebase of Biomolecules Induced by Ultraviolet Radiation Exposure
- PMID: 39866619
- PMCID: PMC11755174
- DOI: 10.1021/acsomega.4c06708
UVREK: Development and Analysis of an Expression Profile Knowledgebase of Biomolecules Induced by Ultraviolet Radiation Exposure
Abstract
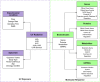
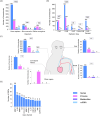
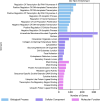
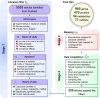
Humans encounter diverse environmental factors which can have impact on their health. One such environmental factor is ultraviolet (UV) radiation, which is part of the physical component of the exposome. UV radiation is the leading cause of skin cancer and is a significant global health concern. A large body of published research has been conducted to uncover the mechanisms underlying the adverse outcomes of UV radiation exposure on living beings. These studies involve identifying the biomolecules induced upon UV radiation exposure. A few previous efforts have attempted to compile this information in the form of a database, but such earlier efforts have certain limitations. To fill this gap, we present a structured database named UVREK (UltraViolet Radiation Expression Knowledgebase), containing manually curated data on biomolecules induced by UV radiation exposure from the published literature. UVREK has compiled information on 985 genes, 470 proteins, 54 metabolites, and 77 miRNAs along with their metadata. Thereafter, an enrichment analysis performed on the human gene set of the UVREK database showed the importance of transcription-related processes in UV-related response and enrichment of pathways involved in cancer and aging. While significantly contributing toward characterizing the physical component of the exposome, we expect that the compiled data in UVREK will serve as a valuable resource for the development of better UV protection mechanisms such as UV sensors and sunscreens. Noteworthily, UVREK is the only resource to date compiling varied types of biomolecular responses to UV radiation with the corresponding metadata. UVREK is openly accessible at https://cb.imsc.res.in/uvrek/.
© 2025 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Impact of ultraviolet radiation and exposome on rosacea: Key role of photoprotection in optimizing treatment.J Cosmet Dermatol. 2021 Nov;20(11):3415-3421. doi: 10.1111/jocd.14020. Epub 2021 Mar 4. J Cosmet Dermatol. 2021. PMID: 33626227 Free PMC article. Review.
-
Broad-spectrum sunscreens provide better protection from solar ultraviolet-simulated radiation and natural sunlight-induced immunosuppression in human beings.J Am Acad Dermatol. 2008 May;58(5 Suppl 2):S149-54. doi: 10.1016/j.jaad.2007.04.035. J Am Acad Dermatol. 2008. PMID: 18410801 Review.
-
The human health effects of ozone depletion and interactions with climate change.Photochem Photobiol Sci. 2011 Feb;10(2):199-225. doi: 10.1039/c0pp90044c. Epub 2011 Jan 20. Photochem Photobiol Sci. 2011. PMID: 21253670 Review.
-
A curated knowledgebase on endocrine disrupting chemicals and their biological systems-level perturbations.Sci Total Environ. 2019 Nov 20;692:281-296. doi: 10.1016/j.scitotenv.2019.07.225. Epub 2019 Jul 16. Sci Total Environ. 2019. PMID: 31349169
-
MeFSAT: a curated natural product database specific to secondary metabolites of medicinal fungi.RSC Adv. 2021 Jan 12;11(5):2596-2607. doi: 10.1039/d0ra10322e. eCollection 2021 Jan 11. RSC Adv. 2021. PMID: 35424258 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
