Meta-omics reveals role of photosynthesis in microbially induced carbonate precipitation at a CO2-rich geyser
- PMID: 39866677
- PMCID: PMC11760937
- DOI: 10.1093/ismeco/ycae139
Meta-omics reveals role of photosynthesis in microbially induced carbonate precipitation at a CO2-rich geyser
Abstract
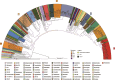
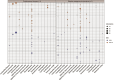
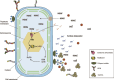
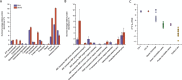
Microbially induced carbonate precipitation (MICP) is a natural process with potential biotechnological applications to address both carbon sequestration and sustainable construction needs. However, our understanding of the microbial processes involved in MICP is limited to a few well-researched pathways such as ureolytic hydrolysis. To expand our knowledge of MICP, we conducted an omics-based study on sedimentary communities from travertine around the CO2-driven Crystal Geyser near Green River, Utah. Using metagenomics and metaproteomics, we identified the community members and potential metabolic pathways involved in MICP. We found variations in microbial community composition between the two sites we sampled, but Rhodobacterales were consistently the most abundant order, including both chemoheterotrophs and anoxygenic phototrophs. We also identified several highly abundant genera of Cyanobacteriales. The dominance of these community members across both sites and the abundant presence of photosynthesis-related proteins suggest that photosynthesis could play a role in MICP at Crystal Geyser. We also found abundant bacterial proteins involved in phosphorous starvation response at both sites suggesting that P-limitation shapes both composition and function of the microbial community driving MICP.
Keywords: MICP; mass spectrometry; metagenomics; metaproteomics; nutrient limitation; rubisco.
© The Author(s) 2024. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Hans WK. The composition of the continental crust. Geochim Cosmochim Acta 1995;59:1217–32. 10.1016/0016-7037(95)00038-2 - DOI
-
- Dupraz C, Visscher PT, Baumgartner LK et al. Microbe-mineral interactions: early carbonate precipitation in a hypersaline lake (Eleuthera Island, Bahamas): microbe-mineral interactions, Eleuthera Island. Bahamas Sedimentology 2004;51:745–65. 10.1111/j.1365-3091.2004.00649.x - DOI
LinkOut - more resources
Full Text Sources

