Computationally Assisted Noncanonical Amino Acid Incorporation
- PMID: 39866708
- PMCID: PMC11758377
- DOI: 10.1021/acscentsci.4c01544
Computationally Assisted Noncanonical Amino Acid Incorporation
Abstract
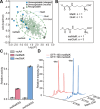
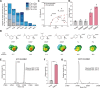
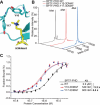
Genetic encoding of noncanonical amino acids (ncAAs) with desired functionalities is an invaluable tool for the study of biological processes and the development of therapeutic drugs. However, existing ncAA incorporation strategies are rather time-consuming and have relatively low success rates. Here, we develop a virtual ncAA screener based on the analysis and modeling of the chemical properties of all reported ncAA substrates to virtually determine the recognition potential of candidate ncAAs. Using this virtual screener, we designed and incorporated several novel Lys and Phe derivatives into proteins for various downstream applications. Among them, the genetic encoding of an electron-rich Phe analog, 3-dimethylamino-phenylalanine, was successfully applied to enhance the cation-π interaction between histone methylation and its reader proteins. Thus, our virtual screener provides a fast and powerful strategy to efficiently incorporate ncAAs with diverse functionalities.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Broadening the Toolkit for Quantitatively Evaluating Noncanonical Amino Acid Incorporation in Yeast.ACS Synth Biol. 2021 Nov 19;10(11):3094-3104. doi: 10.1021/acssynbio.1c00370. Epub 2021 Nov 3. ACS Synth Biol. 2021. PMID: 34730946 Free PMC article.
-
An evolved pyrrolysyl-tRNA synthetase with polysubstrate specificity expands the toolbox for engineering enzymes with incorporation of noncanonical amino acids.Bioresour Bioprocess. 2023 Dec 11;10(1):92. doi: 10.1186/s40643-023-00712-w. Bioresour Bioprocess. 2023. PMID: 38647798 Free PMC article.
-
Genome-Wide Screen for Enhanced Noncanonical Amino Acid Incorporation in Yeast.ACS Synth Biol. 2022 Nov 18;11(11):3669-3680. doi: 10.1021/acssynbio.2c00267. Epub 2022 Nov 8. ACS Synth Biol. 2022. PMID: 36346914 Free PMC article.
-
Future prospects for noncanonical amino acids in biological therapeutics.Curr Opin Biotechnol. 2019 Dec;60:168-178. doi: 10.1016/j.copbio.2019.02.020. Epub 2019 Apr 8. Curr Opin Biotechnol. 2019. PMID: 30974337 Free PMC article. Review.
-
Noncanonical Amino Acid Incorporation in Animals and Animal Cells.Chem Rev. 2024 Nov 27;124(22):12463-12497. doi: 10.1021/acs.chemrev.3c00955. Epub 2024 Nov 14. Chem Rev. 2024. PMID: 39541258 Review.
References
-
- Wang Y.; Zhang J.; Han B.; Tan L.; Cai W.; Li Y.; Su Y.; Yu Y.; Wang X.; Duan X.; Wang H.; Shi X.; Wang J.; Yang X.; Liu T. Noncanonical amino acids as doubly bio-orthogonal handles for one-pot preparation of protein multiconjugates. Nat. Commun. 2023, 14, 974.10.1038/s41467-023-36658-y. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
