Structure Characterization of a Disordered Peptide Using In-Droplet Hydrogen/Deuterium Exchange Mass Spectrometry and Molecular Dynamics
- PMID: 39867440
- PMCID: PMC11758492
- DOI: 10.1021/acsphyschemau.4c00048
Structure Characterization of a Disordered Peptide Using In-Droplet Hydrogen/Deuterium Exchange Mass Spectrometry and Molecular Dynamics
Abstract
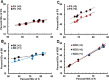
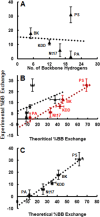
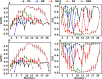
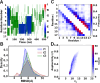
In-droplet hydrogen/deuterium exchange (HDX)-mass spectrometry (MS) experiments have been conducted for peptides of highly varied conformational type. A new model is presented that combines the use of protection factors (PF) from molecular dynamics (MD) simulations with intrinsic HDX rates (k int) to obtain a structure-to-reactivity calibration curve. Using the model, the relationship of peptide structural flexibility and HDX reactivity for different peptides is elucidated. Additionally, the model is used to describe the degree of conformational flexibility and structural bias for the disease-relevant Nt17 peptide; although highly flexible, intrinsically primed for facile conversion to α-helical conformation upon binding with molecular partners imparts significant in-droplet HDX protection for this peptide. In the future, a scale may be developed whereby HDX reactivity is predictive of the degree of structural flexibility and bias (propensity to form 2° structural elements such as α-helix, β-sheet, and β-turn) for intrinsically disordered regions (IDRs). Such structural resolution may ultimately be used for high-throughput screening of IDR structural transformation(s) upon binding of ligands such as drug candidates.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare the following competing financial interest(s): Peng Li and Stephen Valentine have formed the company, Invibragen, Inc. to commercialize VSSI technology.
Figures
Similar articles
-
Recalibrating Protection Factors Using Millisecond Hydrogen/Deuterium Exchange Mass Spectrometry.Anal Chem. 2025 Feb 11;97(5):2648-2657. doi: 10.1021/acs.analchem.4c03631. Epub 2025 Jan 29. Anal Chem. 2025. PMID: 39879324 Free PMC article.
-
In-droplet hydrogen-deuterium exchange to examine protein/peptide solution conformer heterogeneity.Rapid Commun Mass Spectrom. 2023 Aug 30;37(16):e9593. doi: 10.1002/rcm.9593. Rapid Commun Mass Spectrom. 2023. PMID: 37430450 Free PMC article.
-
Developments in rapid hydrogen-deuterium exchange methods.Essays Biochem. 2023 Mar 29;67(2):165-174. doi: 10.1042/EBC20220174. Essays Biochem. 2023. PMID: 36636941 Review.
-
Investigating how intrinsically disordered regions contribute to protein function using HDX-MS.Biochem Soc Trans. 2022 Dec 16;50(6):1607-1617. doi: 10.1042/BST20220206. Biochem Soc Trans. 2022. PMID: 36454645 Review.
-
Bridging protein structure, dynamics, and function using hydrogen/deuterium-exchange mass spectrometry.Protein Sci. 2020 Apr;29(4):843-855. doi: 10.1002/pro.3790. Epub 2019 Nov 25. Protein Sci. 2020. PMID: 31721348 Free PMC article. Review.
Cited by
-
Accessing Different Protein Conformer Ensembles with Tunable Capillary Vibrating Sharp-Edge Spray Ionization.J Phys Chem B. 2025 Feb 6;129(5):1626-1639. doi: 10.1021/acs.jpcb.4c04842. Epub 2025 Jan 29. J Phys Chem B. 2025. PMID: 39878076 Free PMC article.
References
-
- Bradbury E. M.; Danby S. E.; Rattle H. W. E.; Giancotti V. Studies on the Role and Mode of Operation of the Very-Lysine-Rich Histone H1 (F1) in Eukaryote Chromatin. Histone H1 in Chromatin and in H1 . DNA Complexes. Eur. J. Biochem. 1975, 57 (1), 97–105. 10.1111/j.1432-1033.1975.tb02280.x. - DOI - PubMed
-
- Bode W.; Schwager P.; Huber R. The transition of bovine trypsinogen to a trypsin-like state upon strong ligand binding: the refined crystal structures of the bovine trypsinogen-pancreatic trypsin inhibitor complex and of its ternary complex with Ile-Val at 1.9 Å resolution. J. Mol. Biol. 1978, 118 (1), 99.10.1016/0022-2836(78)90246-2. - DOI - PubMed
-
- Dunker A. K.; Babu M. M.; Barbar E.; Blackledge M.; Bondos S. E.; Dosztányi Z.; Dyson H. J.; Forman-Kay J.; Fuxreiter M.; Gsponer J.; et al. What’s in a name? Why these proteins are intrinsically disordered: Why these proteins are intrinsically disordered. Intrinsically Disord. Proteins 2013, 1 (1), e2415710.4161/idp.24157. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
