Molecular Epidemiology of Type F Clostridium perfringens Among Diarrheal Patients and Virulence-Resistance Dynamics - 11 Provinces, China, 2024
- PMID: 39867821
- PMCID: PMC11757904
- DOI: 10.46234/ccdcw2025.013
Molecular Epidemiology of Type F Clostridium perfringens Among Diarrheal Patients and Virulence-Resistance Dynamics - 11 Provinces, China, 2024
Abstract
Introduction: Type F Clostridium perfringens (C. perfringens) represents a significant pathogen in human gastrointestinal diseases, primarily through its cpe gene encoding C. perfringens enterotoxin (CPE). This investigation examined the prevalence, antimicrobial resistance patterns, and genetic characteristics of Type F C. perfringens within the Chinese population.
Methods: The study analyzed 2,068 stool samples collected from 11 provincial hospitals in 2024. Antimicrobial susceptibility testing was conducted following Clinical & Laboratory Standards Institute (CLSI) guidelines, while whole-genome sequencing provided detailed genetic profiles. Evolutionary relationships and clonal transmission patterns were investigated through phylogenetic and genetic environment analyses.
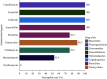
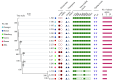
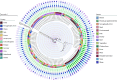
Results: The prevalence of Type F C. perfringens was 2.38%, with isolates predominantly identified in human clinical samples and higher detection rates in gastroenterology departments. Notably, 47.1% of isolates demonstrated high resistance to metronidazole, while all exhibited intermediate resistance to erythromycin. Phylogenetic analysis revealed high similarity among isolates from patients within the same province (single-nucleotide polymorphism (SNPs)<100), and genetic environment analysis indicated potential horizontal gene transfer between animal and human strains.
Conclusions: This investigation predominantly identified Type F C. perfringens in human clinical cases, with sporadic detection in pets and food products. These findings highlight the emergence of Type F C. perfringens outbreaks among diarrheal patients, emphasizing the necessity for targeted interventions as virulence factors increase.
Keywords: Type F Clostridium perfringens; antimicrobial resistance; diarrheal patients; virulence factors.
Copyright © 2025 by Chinese Center for Disease Control and Prevention.
Conflict of interest statement
No conflicts of interest.
Figures

References
-
- Kiu R, Caim S, Painset A, Pickard D, Swift C, Dougan G, et al Phylogenomic analysis of gastroenteritis-associated Clostridium perfringens in England and Wales over a 7-year period indicates distribution of clonal toxigenic strains in multiple outbreaks and extensive involvement of enterotoxin-encoding (CPE) plasmids. Microb Genom. 2019;5(10):e000297. doi: 10.1099/mgen.0.000297. - DOI - PMC - PubMed
-
- Zhao FL, Gao X, Zhen BJ, Zhang P, Luo YX, Wang RP, et al Laboratory detection and analysis of a suspected diarrhea outbreak caused by Clostridium perfringens. Chin J Food Hyg. 2023;35(7):1109–13. doi: 10.13590/j.cjfh.2023.07.021. - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
