Loss of Parp7 increases type I interferon signalling and reduces pancreatic tumour growth by enhancing immune cell infiltration
- PMID: 39867896
- PMCID: PMC11759301
- DOI: 10.3389/fimmu.2024.1513595
Loss of Parp7 increases type I interferon signalling and reduces pancreatic tumour growth by enhancing immune cell infiltration
Abstract
Background: Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal forms of cancer, and despite low incidence rates, it remains the sixth leading cause of cancer related deaths worldwide. Immunotherapy, which aims to enhance the immune system's ability to recognize and eliminate cancer cells, has emerged as a promising approach in the battle against PDAC. PARP7, a mono-ADP-ribosyltransferase, is a negative regulator of the type I interferon (IFN-I) pathway and has been reported to reduce anti-tumour immunity.
Methods: We used murine pancreatic cancer cells, CR705, CRISPR/Cas9, in vivo tumour models and spectral flow cytometry to determine the role of PARP7 in pancreatic tumour growth.
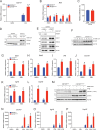
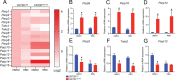
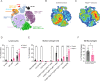
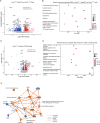
Results: Loss of Parp7 elevated the levels of interferon stimulated gene factor 3 (ISGF3) and its downstream target genes, even in the absence of STING. Cancer cells knocked out for Parp7 (CR705Parp7KO) produced smaller tumours than control cells (CR705Cas9) when injected into immunocompetent mice. Transcriptomic analyses revealed that CR705Parp7KO tumours had increased expression of genes involved in immunoregulatory interactions and interferon signalling pathways. Characterization of tumour infiltrating leukocyte (TIL) populations showed that CR705Parp7KO tumours had higher proportions of natural killer cells, CD8+ T cells and a lower proportion of anti-inflammatory macrophages (M2). The overall TIL profile of CR705Parp7KO tumours was suggestive of a less suppressive microenvironment.
Conclusions: Our data show that loss of Parp7 reduces PDAC tumour growth by increasing the infiltration of immune cells and enhancing anti-tumour immunity. These findings provide support to pursue PARP7 as a therapeutic target for cancer treatment.
Keywords: CRISPR/Cas9; Poly-ADP-ribose polymerase 7; pancreatic cancer; tumour infiltrating leukocytes; type I interferon.
Copyright © 2025 Kannen, Rasmussen, Das, Giuliana, Izzati, Choksi, Erlingsson, Olafsen, Åhrling, Cappello, Teino, Maimets, Jaudzems, Gulbinas, Dambrauskas, Edgar, Grant and Matthews.
Conflict of interest statement
JM was a consultant for Duke Street Bio Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

