Nationwide multi-centric prospective study for the identification of biomarkers to predict the treatment responses of nivolumab through comprehensive analyses of pretreatment plasma exosome mRNAs from head and neck cancer patients (BIONEXT study)
- PMID: 39867897
- PMCID: PMC11758179
- DOI: 10.3389/fimmu.2024.1464419
Nationwide multi-centric prospective study for the identification of biomarkers to predict the treatment responses of nivolumab through comprehensive analyses of pretreatment plasma exosome mRNAs from head and neck cancer patients (BIONEXT study)
Abstract
Background: Nivolumab paved a new way in the treatment of patients with recurrent or metastatic (RM) head and neck squamous cell carcinoma (RM-HNSCC). However, the limited rates of long-term survivors (< 20%) demand a robust prognostic biomarker. This nationwide multi-centric prospective study aimed to identify a plasma exosome (PEX) mRNA signature, which serves as a companion diagnostic of nivolumab and provides a biological clue to develop effective therapies for a majority of non-survivors.
Methods: Pre-treatment plasmas (N = 104) of RM-HNSCC patients were subjected to comprehensive PEX mRNA analyses for prognostic marker discovery and validation. In parallel, paired treatment-naïve tumor and plasma samples (N = 20) were assayed to elucidate biological implications of the PEX mRNA signature.
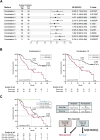
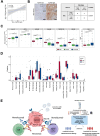
Results: Assays for pre-treatment blood samples (N = 104) demonstrated that a combination of 6 candidate PEX mRNAs plus neutrophil-to-lymphocyte ratio precisely distinguished non-survivors from >2-year survivors (2-year OS; 0% vs 57.7%; P = 0.000124) with a high hazard ratio of 2.878 (95% CI 1.639-5.055; P = 0.0002348). Parallel biological assays demonstrated that in the paired treatment-naïve HNSCC tumor and plasma samples (N = 20), PEX HLA-E mRNA (a non-survivor-predicting marker) was positively corelated with overexpression of HLA-E protein (P = 0.0191) and the dense population of tumor-infiltrating NK cells (P = 0.024) in the corresponding tumor, suggesting that the HLA-E-NKG2A immune checkpoint may inhibit the antitumor effect of PD-1blockade.
Conclusion: The PEX mRNA signature could be useful as a companion diagnostic of nivolumab. The combination of an anti-NKG2A antibody (i.e., monalizumab) and nivolumab may serve as a treatment option for non-survivors predicted by a RT-qPCR-based pre-treatment measurement of PEX mRNAs.
Keywords: HLA-E; biomarker; exosome; head and neck cancer; nivolumab.
Copyright © 2025 Sato, Toh, Murakami, Nakano, Hongo, Matsuo, Hashimoto, Sugasawa, Yamazaki, Ueki, Nakashima, Uryu, Ono, Umeno, Ueda, Kano, Tsukahara, Watanabe, Ota, Monden, Iwae, Maruo, Asada, Hanai, Sano, Ozawa, Asakage, Fukusumi and Masuda.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Ferris RL, Blumenschein G, Jr., Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab vs investigator's choice in recurrent or metastatic squamous cell carcinoma of the head and neck: 2-year long-term survival update of CheckMate 141 with analyses by tumor PD-L1 expression. Oral Oncol. (2018) 81:45–51. doi: 10.1016/j.oraloncology.2018.04.008 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

