Interferon activation in bone marrow long-lived plasma cells in systemic lupus erythematosus
- PMID: 39867907
- PMCID: PMC11757124
- DOI: 10.3389/fimmu.2024.1499551
Interferon activation in bone marrow long-lived plasma cells in systemic lupus erythematosus
Abstract
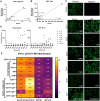
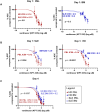
While durable antibody responses from long-lived plasma cell (LLPC) populations are important for protection against pathogens, LLPC may be harmful if they produce antibodies against self-proteins or self-nuclear antigens as occurs in autoimmune diseases such as systemic lupus erythematosus (SLE). Thus, the elimination of autoreactive LLPC may improve the treatment of antibody-driven autoimmune diseases. However, LLPC remain a challenging therapeutic target. Here, we compare the matched bone marrow (BM) and peripheral blood (PBL) plasma cell (PC) compartments of SLE and healthy donors (HD). We show a similar distribution of CD138- and CD138+ PC, including putative LLPC (CD19- CD138+ CD38+), between SLE and HD BM. For both SLE and HD, CD138+ PC are at a higher frequency in BM than PBL. Expression of Ki-67 associates with the PBL compartment where it is found on all PC subsets regardless of CD19 or CD138 expression. Transcriptomic analysis identifies an interferon (IFN) gene signature in transitional B cells in the SLE BM, but surprisingly also in the BM PC derived from SLE. BM PC and B cells phosphorylate STAT1 in response to type I IFN stimulation in vitro, but with decreased fold change compared to those from the PBL. While BM PC bind type I IFN receptor-blocking antibody anifrolumab, it is to a lesser degree than circulating B cells. Anti-nuclear autoantibodies (ANA) are found in the BM supernatant and PBL serum of SLE patients. Both SLE and HD BM-derived PC have increased survival compared to their PBL counterparts when treated with verdinexor. In summary, these findings show evidence of IFN activation in BM PC from SLE.
Keywords: B cell; anifrolumab; bone marrow; interferon; lupus (SLE); plasma cell (PC); transitional B cell; verdinexor.
Copyright © 2025 Alzamareh, Meednu, Nandedkar-Kulkarni, Krenitsky, Barnard, Yasaka, Durrett, Thakar, Rangel-Moreno, Anolik and Barnas.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures
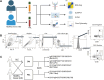
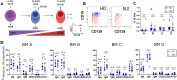



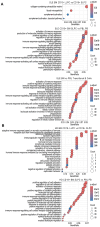
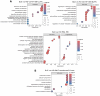


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

