This is a preprint.
Conservation of symmetry breaking at the level of chromatin accessibility between fly species with unrelated anterior determinants
- PMID: 39868093
- PMCID: PMC11760685
- DOI: 10.1101/2025.01.13.632851
Conservation of symmetry breaking at the level of chromatin accessibility between fly species with unrelated anterior determinants
Abstract
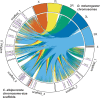
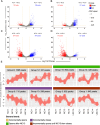
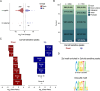
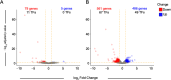
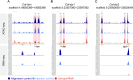
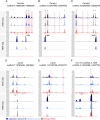
Establishing the anterior-posterior body axis is a fundamental process during embryogenesis, and the fruit fly, Drosophila melanogaster, provides one of the best-known case studies of this process. In Drosophila, localized mRNA of bicoid serves as anterior determinant (AD). Bicoid engages in a concentration-dependent competition with nucleosomes and initiates symmetry-breaking along the AP axis by promoting chromatin accessibility at the loci of transcription factor (TF) genes that are expressed in the anterior of the embryo. However, ADs of other fly species are unrelated and structurally distinct, and little is known about how they function. We addressed this question in the moth fly, Clogmia albipunctata, in which a maternally expressed transcript isoform of the pair-rule segmentation gene odd-paired is localized in the anterior egg and has been co-opted as AD. We provide a de novo assembly and annotation of the Clogmia genome and describe how knockdown of zelda and maternal odd-paired transcript affect chromatin accessibility and expression of TF-encoding loci. The results of these experiments suggest direct roles of Cal-Zld in opening and closing chromatin during nuclear cleavage cycles and show that Clogmia's maternal odd-paired activity promotes chromatin accessibility and anterior expression during the early phase of zygotic genome activation at Clogmia's homeobrain and sloppy-paired loci. We conclude that unrelated dipteran ADs initiate anterior-posterior axis-specification at the level of enhancer accessibility and that homeobrain and sloppy-paired homologs may serve a more widely conserved role in the initiation of anterior pattern formation given their early anterior expression and function in head development in other insects.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures


Similar articles
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
T-bet expressing Tr1 cells driven by dietary signals dominate the small intestinal immune landscape.bioRxiv [Preprint]. 2025 Jul 4:2025.06.30.662190. doi: 10.1101/2025.06.30.662190. bioRxiv. 2025. PMID: 40747421 Free PMC article. Preprint.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Characterization Eclosion Hormone Receptor function reveals differential hormonal control of ecdysis during Drosophila development.bioRxiv [Preprint]. 2025 May 4:2025.04.03.646974. doi: 10.1101/2025.04.03.646974. bioRxiv. 2025. PMID: 40568096 Free PMC article. Preprint.
-
Incentives for preventing smoking in children and adolescents.Cochrane Database Syst Rev. 2012 Oct 17;10:CD008645. doi: 10.1002/14651858.CD008645.pub2. Cochrane Database Syst Rev. 2012. Update in: Cochrane Database Syst Rev. 2017 Jun 06;6:CD008645. doi: 10.1002/14651858.CD008645.pub3. PMID: 23076949 Updated.
References
-
- Driever W, Nüsslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988;54(1):95–104. - PubMed
-
- Driever W, Nüsslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988;54(1):83–93. - PubMed
-
- St Johnston D, Nüsslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 1992;68(2):201–19. - PubMed
-
- Surkova S, Golubkova E, Mamon L, Samsonova M. Dynamic maternal gradients and morphogenetic networks in Drosophila early embryo. Bio Systems. 2018;173:207–13. - PubMed
-
- Ma J, He F, Xie G, Deng GX. Maternal AP determination in the Drosophila oocyte and embryo. Wiley Interdiscip Rev Dev Biol. 2016;5:562–81. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
