This is a preprint.
Identification of novel Kv1.3 channel-interacting proteins using proximity labelling in T-cells
- PMID: 39868101
- PMCID: PMC11760797
- DOI: 10.1101/2025.01.16.633279
Identification of novel Kv1.3 channel-interacting proteins using proximity labelling in T-cells
Abstract
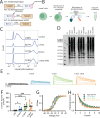
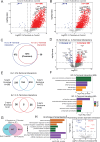
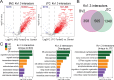
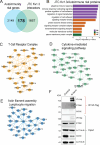
Potassium channels regulate membrane potential, calcium flux, cellular activation and effector functions of adaptive and innate immune cells. The voltage-activated Kv1.3 channel is an important regulator of T cell-mediated autoimmunity and microglia-mediated neuroinflammation. Kv1.3 channels, via protein-protein interactions, are localized with key immune proteins and pathways, enabling functional coupling between K+ efflux and immune mechanisms. To gain insights into proteins and pathways that interact with Kv1.3 channels, we applied a proximity-labeling proteomics approach to characterize protein interactors of the Kv1.3 channel in activated T-cells. Biotin ligase TurboID was fused to either N or C termini of Kv1.3, stably expressed in Jurkat T cells and biotinylated proteins in proximity to Kv1.3 were enriched and quantified by mass spectrometry. We identified over 1,800 Kv1.3 interactors including known interactors (beta-integrins, Stat1) although majority were novel. We found that the N-terminus of Kv1.3 preferentially interacts with protein synthesis and protein trafficking machinery, while the C-terminus interacts with immune signaling and cell junction proteins. T-cell Kv1.3 interactors included 335 cell surface, T-cell receptor complex, mitochondrial, calcium and cytokine-mediated signaling pathway and lymphocyte migration proteins. 178 Kv1.3 interactors in T-cells also represent genetic risk factors of T cell-mediated autoimmunity, including STIM1, which was further validated using co-immunoprecipitation. Our studies reveal novel proteins and molecular pathways that interact with Kv1.3 channels in adaptive (T-cell) and innate immune (microglia), providing a foundation for how Kv1.3 channels may regulate immune mechanisms in autoimmune and neurological diseases.
Keywords: Potassium channel; T cell; autoimmune disease; interactions; proteomics; proximity labeling.
Conflict of interest statement
STATEMENTS Statement of Ethics The authors have no ethical conflicts to disclose. Disclosure Statement The authors have no conflicts of interest to declare.
Figures


References
-
- He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433(7028):826–33. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
