This is a preprint.
TEAD switches interacting partners along neural progenitor lineage progression to execute distinct functions
- PMID: 39868115
- PMCID: PMC11760702
- DOI: 10.1101/2024.12.19.629472
TEAD switches interacting partners along neural progenitor lineage progression to execute distinct functions
Update in
-
TEAD switches interacting partners along neural progenitor lineage progression to execute distinct functions.Genes Dev. 2025 Jul 1;39(13-14):849-867. doi: 10.1101/gad.352632.125. Genes Dev. 2025. PMID: 40389325
Abstract
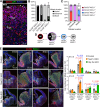
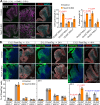
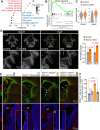
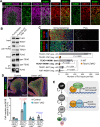
The TEAD family of transcription factors are best known as the DNA-binding factor in the Hippo pathway, where they act by interacting with transcriptional coactivators YAP and TAZ (YAP/TAZ). Despite the importance of the Hippo pathway, the in vivo functions of TEAD in mammals have not been well established. By comparing mouse mutants lacking TEAD1 and TEAD2 (TEAD1/2) to those lacking YAP/TAZ, we found that TEAD1/2 have both YAP/TAZ-dependent and -independent functions during ventral telencephalon development. TEAD1/2 loss and YAP/TAZ loss similarly disrupt neuroepithelial apical junctions. However, the impacts of their losses on progenitor lineage progression are essentially opposite: Whereas YAP/TAZ loss depletes early progenitors and increases later progenitors-consistent with their established function in promoting progenitor self-renewal and proliferation, TEAD1/2 loss expands early progenitors and reduces late progenitors, indicating that TEAD1/2 promote lineage progression. We further show that TEAD1/2 promote neural progenitor lineage progression by, at least in part, inhibiting Notch signaling and by cooperating with Insulinoma-associated 1 (INSM1). Orthologs of TEAD and INSM1 have been shown to cooperatively regulate neuronal cell fate decisions in worms and flies. Our study reveals a remarkable evolutionary conservation of the function of this transcription factor complex during metazoan neural development.
Keywords: VGLL4; apical progenitor; basal ganglia; basal progenitor; intermediate progenitor; interneuron; medial ganglionic eminence (MGE); neurogenesis; radial glia; subapical progenitor.
Conflict of interest statement
Competing Interest Statement The authors declare no competing interests.
Figures



References
-
- Allen Institute for Brain Science. Allen Brain Atlas Developing Mouse Brain [dataset].
-
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. 1997. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science 278: 474–476. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
