This is a preprint.
Microbiota assembly of specific pathogen-free neonatal mice
- PMID: 39868118
- PMCID: PMC11761686
- DOI: 10.1101/2025.01.14.633035
Microbiota assembly of specific pathogen-free neonatal mice
Update in
-
Bacterial community assembly of specific pathogen-free neonatal mice.Microbiome. 2025 Feb 7;13(1):46. doi: 10.1186/s40168-025-02043-8. Microbiome. 2025. PMID: 39920864 Free PMC article.
Abstract
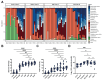
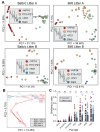
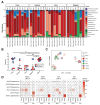
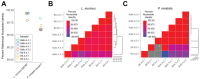
Background: Neonatal mice are frequently used to model diseases that affect human infants. Microbial community composition has been shown to impact disease progression in these models. Despite this, the maturation of the early-life murine microbiome has not been well-characterized. We address this gap by characterizing the assembly of the bacterial microbiota of C57BL/6 and BALB/c litters from birth to adulthood across multiple independent litters.
Results: The fecal microbiome of young pups is simple, dominated by only a few pioneering bacterial taxa. These taxa are present at low levels in the microbiota of multiple maternal body sites, precluding a clear identification of maternal source. The pup microbiota begins diversifying after fourteen days, coinciding with the beginning of coprophagy and the consumption of solid foods. Pup stool bacterial community composition and diversity are not significantly different from dams from day 21 onwards. Short-read shotgun sequencing-based metagenomic profiling of young pups enabled the assembly of metagenome-assembled genomes for strain-level analysis of these pioneer Ligilactobacillus, Streptococcus, and Proteus species.
Conclusions: Assembly of the murine microbiome occurs over the first weeks of postnatal life and is largely complete by day 21. This detailed view of bacterial community development across multiple commonly employed mouse strains informs experimental design, allowing researchers to better target interventions before, during, or after the maturation of the bacterial microbiota. The source of pioneer bacterial strains appears heterogeneous, as the most abundant taxa identified in young pup stool were found at low levels across multiple maternal body sites, suggesting diverse routes for seeding of the murine microbiome.
Keywords: development; early-life; microbiome; microbiota; mother-infant transmission; neonatal; pioneer species; seeding.
Conflict of interest statement
Competing interests We confirm that this manuscript has not been published elsewhere and is not under consideration by another journal. All authors have approved the manuscript and agree with its submission to Microbiome. There are no conflicts of interest to report, and the care of animals adhered to institutional guidelines at Washington University School of Medicine.
Figures
Similar articles
-
Bacterial community assembly of specific pathogen-free neonatal mice.Microbiome. 2025 Feb 7;13(1):46. doi: 10.1186/s40168-025-02043-8. Microbiome. 2025. PMID: 39920864 Free PMC article.
-
Intestinal microbiota domination under extreme selective pressures characterized by metagenomic read cloud sequencing and assembly.BMC Bioinformatics. 2019 Dec 2;20(Suppl 16):585. doi: 10.1186/s12859-019-3073-1. BMC Bioinformatics. 2019. PMID: 31787070 Free PMC article.
-
Birth Mode Does Not Determine the Presence of Shared Bacterial Strains between the Maternal Vaginal Microbiome and the Infant Stool Microbiome.Microbiol Spectr. 2023 Aug 17;11(4):e0061423. doi: 10.1128/spectrum.00614-23. Epub 2023 Jun 20. Microbiol Spectr. 2023. PMID: 37338388 Free PMC article.
-
Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery.Nat Med. 2017 Mar;23(3):314-326. doi: 10.1038/nm.4272. Epub 2017 Jan 23. Nat Med. 2017. PMID: 28112736 Free PMC article.
-
Microbial programming of health and disease starts during fetal life.Birth Defects Res C Embryo Today. 2015 Dec;105(4):265-77. doi: 10.1002/bdrc.21117. Epub 2015 Dec 10. Birth Defects Res C Embryo Today. 2015. PMID: 26663884 Review.
References
-
- Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host and Microbe. 2015;17:690–703. - PubMed
-
- Roswall J, Olsson LM, Kovatcheva-Datchary P, Nilsson S, Tremaroli V, Simon M-C, et al. Developmental trajectory of the healthy human gut microbiota during the first 5 years of life. Cell host & microbe. 2021;29:765–776.e3. - PubMed
-
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11971–5. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
