This is a preprint.
Systematic Determination of the Impact of Structural Edits on Peptide Accumulation into Mycobacteria
- PMID: 39868157
- PMCID: PMC11760776
- DOI: 10.1101/2025.01.17.633618
Systematic Determination of the Impact of Structural Edits on Peptide Accumulation into Mycobacteria
Update in
-
Systematic Determination of the Impact of Structural Edits on Peptide Accumulation into Mycobacteria.ACS Chem Biol. 2025 Aug 15;20(8):1962-1979. doi: 10.1021/acschembio.5c00330. Epub 2025 Aug 1. ACS Chem Biol. 2025. PMID: 40748788 Free PMC article.
Abstract
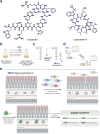
Understanding the factors that influence the accumulation of molecules beyond the mycomembrane of Mycobacterium tuberculosis (Mtb) - the main barrier to accumulation - is essential for developing effective antimycobacterial agents. In this study, we investigated two design principles commonly observed in natural products and mammalian cell-permeable peptides: backbone N-alkylation and macrocyclization. To assess how these structural edits impact molecule accumulation beyond the mycomembrane, we utilized our recently developed Peptidoglycan Accessibility Click-Mediated Assessment (PAC-MAN) assay for live-cell analysis. Our findings provide the first empirical evidence that peptide macrocyclization generally enhances accumulation in mycobacteria, while N-alkylation influences accumulation in a context-dependent manner. We examined these design principles in the context of two peptide antibiotics, tridecaptin A1 and griselimycin, which revealed the roles of N-alkylation and macrocyclization in improving both accumulation and antimicrobial activity against mycobacteria in specific contexts. Together, we present a working model for strategic structural modifications aimed at enhancing the accumulation of molecules past the mycomembrane. More broadly, our results also challenge the prevailing belief in the field that large and hydrophilic molecules, such as peptides, cannot readily traverse the mycomembrane.
Figures





References
-
- Global Tuberculosis Report 2023 - TB Incidence. https://www.who.int/teams/global-tuberculosis-programme/tb-reports/globa... (accessed 2024-06-03).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
