This is a preprint.
Identification of a minimal Alu domain required for retrotransposition
- PMID: 39868163
- PMCID: PMC11760393
- DOI: 10.1101/2024.12.16.628748
Identification of a minimal Alu domain required for retrotransposition
Update in
-
Identification of a minimal Alu domain required for retrotransposition.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf526. doi: 10.1093/nar/gkaf526. Nucleic Acids Res. 2025. PMID: 40568936 Free PMC article.
Abstract
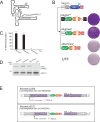
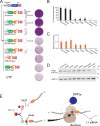
Alu elements are primate-specific retrotransposon sequences that comprise ~11% of human genomic DNA. Alu sequences contain an internal RNA polymerase III promoter and the resultant Alu RNA transcripts mobilize by a replicative process termed retrotransposition. Alu retrotransposition requires the Long INterspersed Element-1 (LINE-1) open reading frame 2-encoded protein (ORF2p). Current models propose that Alu RNA binds to signal recognition particle proteins 9 and 14 (SRP9/14) and localizes to ribosomes, which allows Alu to 'hijack' L1 ORF2p. Here, we used HeLa cell-based retrotransposition assays to define a minimal Alu domain necessary for retrotransposition. We demonstrate that Alu transcripts expressed from a cytomegalovirus (CMV) RNA polymerase II promoter can efficiently undergo retrotransposition. The use of an external CMV promoter to express Alu RNA allowed us to construct separation-of-function mutations to examine the effects of large deletions within the Alu sequence on retrotransposition. Deletion mutagenesis demonstrated that a 46 nucleotide (nt) domain located at the 5' end of the Alu RNA transcript is necessary for Alu retrotransposition. Consistent with current models, the 46 nt 5' Alu domain associates with SRP9/14 in HeLa-HA cell extracts and can promote a single round of retrotransposition in HeLa-HA cells. We propose that the 46 nt 5' Alu domain forms a discrete structure that allows for SRP 9/14 binding and ribosomal association, thereby allowing the Alu poly(A) tract to compete with the L1 poly(A) tail for ORF2p RNA binding to mediate its retrotransposition.
Conflict of interest statement
COMPETING INTERESTS J.V.M. is an inventor on patent US6150160, is a paid consultant for Gilead Sciences, serves on the scientific advisory board of Tessera Therapeutics Inc. (where he is paid as a consultant and has equity options), and has licensed reagents to Merck Pharmaceutical. He also recently served on the American Society of Human Genetics Board of Directors. The other authors do not declare competing interests.
Figures


References
-
- Ullu E. and Tschudi C. (1984) Alu sequences are processed 7SL RNA genes. Nature, 312, 171–172. - PubMed
-
- Boeke J.D., Garfinkel D.J., Styles C.A. and Fink G.R. (1985) Ty elements transpose through an RNA intermediate. Cell, 40, 491–500. - PubMed
-
- Dewannieux M., Esnault C. and Heidmann T. (2003) LINE-mediated retrotransposition of marked Alu sequences. Nat Genet, 35, 41–48. - PubMed
-
- Cordaux R., Hedges D.J., Herke S.W. and Batzer M.A. (2006) Estimating the retrotransposition rate of human Alu elements. Gene, 373, 134–137. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
