This is a preprint.
Keratin intermediate filaments mechanically position melanin pigments for genome photoprotection
- PMID: 39868182
- PMCID: PMC11761041
- DOI: 10.1101/2025.01.15.632531
Keratin intermediate filaments mechanically position melanin pigments for genome photoprotection
Abstract
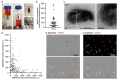
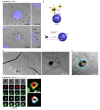
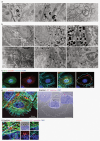
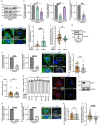
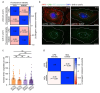
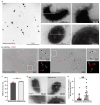
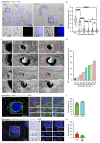
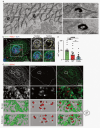
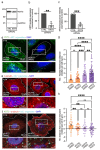
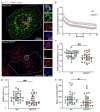
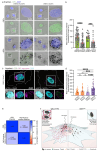
Melanin pigments block genotoxic agents by positioning on the sun-exposed side of human skin keratinocytes' nucleus. How this position is regulated and its role in genome photoprotection remains unknown. By developing a model of human keratinocytes internalizing extracellular melanin into pigment organelles, we show that keratin 5/14 intermediate filaments mechanically control the 3D perinuclear position of pigments, shielding DNA from photodamage. Imaging and microrheology in human disease-related model identify structural keratin cages surrounding pigment organelles to stiffen their microenvironment and maintain their 3D position. Optimum pigment spatialization is required for DNA photoprotection and rely on the interplay between intermediate filaments and microtubules bridged by plectin cytolinkers. Thus, the mechanically-driven proximity of pigment organelles to the nucleus is a key photoprotective parameter. Uncovering how human skin counteracts solar radiation by positioning the melanin microparasol next to the genome anticipates that dynamic spatialization of organelles is a physiological UV stress response.
Keywords: intermediate filaments; keratinocyte; keratins; melanin; microtubules; organelle distribution; skin pigmentation and photoprotection.
Conflict of interest statement
Competing Interest The authors declare a competing interest. Marion Plessis, Charlène Gayrard, Françoise Bernerd and Christine Duval are full time employees of L’Oreal Research and Innovation, which also provided a financial support through a research contract agreement with Structure and Membrane Compartments’s team, Institut Curie, PSL Research University, CNRS, UMR144, 75005 Paris, France.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
