This is a preprint.
Community assembly modeling of microbial evolution within Barrett's esophagus and esophageal adenocarcinoma
- PMID: 39868296
- PMCID: PMC11760701
- DOI: 10.1101/2025.01.14.633020
Community assembly modeling of microbial evolution within Barrett's esophagus and esophageal adenocarcinoma
Abstract
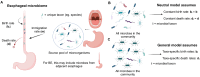
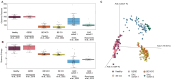
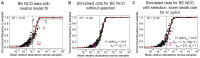
Mathematical modeling of somatic evolution, a process impacting both host cells and microbial communities in the human body, can capture important dynamics driving carcinogenesis. Here we considered models for esophageal adenocarcinoma (EAC), a cancer that has dramatically increased in incidence over the past few decades in Western populations, with high case fatality rates due to late-stage diagnoses. Despite advancements in genomic analyses of the precursor Barrett's esophagus (BE), prevention of late-stage EAC remains a significant clinical challenge. Previous microbiome studies in BE and EAC have focused on quantifying static microbial abundance differences rather than evolutionary dynamics. Using whole genome sequencing data from esophageal tissues, we first applied a robust bioinformatics pipeline to extract non-host DNA reads, mapped these putative reads to microbial taxa, and retained those taxa with high genomic coverage. When applying mathematical models of microbial evolution to sequential stages of progression to EAC, we observed evidence of neutral dynamics in community assembly within normal esophageal tissue and BE, but not EAC. In a case-control study of BE patients who progressed to EAC cancer outcomes (CO) versus those who had non-cancer outcomes (NCO) during follow-up (mean=10.5 years), we found that Helicobacter pylori deviated significantly from the neutral expectation in BE NCO, suggesting that factors related to H. pylori or H. pylori infection itself may influence EAC risk. Additionally, simulations incorporating selection recapitulated non-neutral behaviors observed in the datasets. Formally modeling dynamics during progression holds promise in clinical applications by offering a deeper understanding of microbial involvement in cancer development.
Keywords: Barrett’s esophagus; Esophageal adenocarcinoma; Helicobacter pylori; cancer microbiome; community assembly; mathematical modeling.
Conflict of interest statement
Competing Interests D.M. is a consultant for BiomeSense, Inc., has equity and receives income. The terms of these arrangements have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. LBA is a co-founder, CSO, scientific advisory member, and consultant for io9, has equity and receives income. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. L.B.A. is a compensated member of the scientific advisory board of Inocras. L.B.A.’s spouse is an employee of Hologic, Inc. L.B.A. declares U.S. provisional applications with serial numbers: 63/289,601; 63/269,033; 63/366,392; 63/412,835 as well as international patent application PCT/US2023/010679. L.B.A. is also an inventor of a US Patent 10,776,718 for source identification by non-negative matrix factorization. R.K. is a scientific advisory board member, and consultant for BiomeSense, Inc., has equity and receives income. He is a scientific advisory board member and has equity in GenCirq. He is a consultant and scientific advisory board member for DayTwo, and receives income. He has equity in and acts as a consultant for Cybele. He is a co-founder of Biota, Inc., and has equity. He is a cofounder of Micronoma, and has equity and is a scientific advisory board member. The terms of these arrangements have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. K.C. has research grant support from Phathom Pharmaceuticals.
Figures


Similar articles
-
Surveillance of Barrett's oesophagus: exploring the uncertainty through systematic review, expert workshop and economic modelling.Health Technol Assess. 2006 Mar;10(8):1-142, iii-iv. doi: 10.3310/hta10080. Health Technol Assess. 2006. PMID: 16545207
-
Behavioral interventions to reduce risk for sexual transmission of HIV among men who have sex with men.Cochrane Database Syst Rev. 2008 Jul 16;(3):CD001230. doi: 10.1002/14651858.CD001230.pub2. Cochrane Database Syst Rev. 2008. PMID: 18646068
-
The Effect of Endoscopic Surveillance in Patients With Barrett's Esophagus: A Systematic Review and Meta-analysis.Gastroenterology. 2018 Jun;154(8):2068-2086.e5. doi: 10.1053/j.gastro.2018.02.022. Epub 2018 Feb 16. Gastroenterology. 2018. PMID: 29458154 Free PMC article.
-
Helicobacter pylori infection does not influence the progression from gastroesophageal reflux disease to Barrett's esophagus to esophageal adenocarcinoma.Minerva Gastroenterol (Torino). 2024 Dec;70(4):454-462. doi: 10.23736/S2724-5985.24.03609-X. Epub 2024 May 10. Minerva Gastroenterol (Torino). 2024. PMID: 38727697
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
References
-
- Battaglia TW, Mimpen IL, Traets JJH, van Hoeck A, Zeverijn LJ, Geurts BS, et al. A pan-cancer analysis of the microbiome in metastatic cancer. Cell. 2024;187: 2324–2335.e19. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
