Ring A Cleaving Beta-Diketone Hydrolase Is a Key Enzyme of Steroid Degradation in Anaerobic Bacteria
- PMID: 39868648
- PMCID: PMC11780191
- DOI: 10.1111/1462-2920.70034
Ring A Cleaving Beta-Diketone Hydrolase Is a Key Enzyme of Steroid Degradation in Anaerobic Bacteria
Abstract
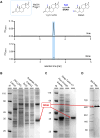
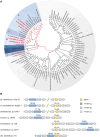
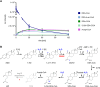
Bacterial degradation of ubiquitous and persistent steroids such as steroid hormones is important for their removal from the environment. Initial studies of steroid degradation in anaerobic bacteria suggested that ring-cleaving hydrolases are involved in oxygen-independent sterane skeleton degradation. However, the enzymes involved in ring A cleavage of the common intermediate androsta-1,4-diene-3,17-dione have remained unknown. Here, we enriched a ring A hydrolase from cholesterol/nitrate grown Sterolibacterium denitrificans and from Escherichia coli after heterologous expression of its gene. This enzyme specifically cleaves the cyclic 1,3-diketone of the central degradation intermediate, androsta-1,3,17-trione to 1,17-dioxo-2,3-seco-androstan-3-oate (DSAO), a hallmark reaction of anaerobic steroid degradation. The highly conserved ring A hydrolase was identified in all known and many previously unknown steroid-degrading proteobacteria. Using enriched enzymes, we enzymatically produced DSAO from the chemically synthesised androsta-1-en-3,17-dione precursor, allowing the identification of subsequent metabolites involved in ring A degradation. The results obtained suggest the involvement of an additional hydrolase, an aldolase, and a β-oxidation-like cascade for complete ring A degradation to form the three-ring 5,10-seco-1,2,3,4-tetranorandrosta-5,17-dione. The results identified a key enzyme of anaerobic steroid degradation that may serve as a functional marker for monitoring steroid contaminant degradation at anoxic environmental sites.
Keywords: beta‐diketon hydrolase; ring hydrolase; sterane skeleton; steroid degradation.
Environmental Microbiology© 2024 The Author(s). Environmental Microbiology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Capyk, J. K. , Casabon I., Gruninger R., Strynadka N. C., and Eltis L. D.. 2011. “Activity of 3‐Ketosteroid 9α‐Hydroxylase (KshAB) Indicates Cholesterol Side Chain and Ring Degradation Occur Simultaneously in Mycobacterium tuberculosis .” Journal of Biological Chemistry 286: 40717–40724. - PMC - PubMed
-
- Chiang, Y.‐R. , Ismail W., Müller M., and Fuchs G.. 2007. “Initial Steps in the Anoxic Metabolism of Cholesterol by the Denitrifying Sterolibacterium denitrificans .” Journal of Biological Chemistry 282: 13240–13249. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

