Pharmacological Dissection Identifies Retatrutide Overcomes the Therapeutic Barrier of Obese TNBC Treatments through Suppressing the Interplay between Glycosylation and Ubiquitylation of YAP
- PMID: 39868848
- PMCID: PMC11923992
- DOI: 10.1002/advs.202407494
Pharmacological Dissection Identifies Retatrutide Overcomes the Therapeutic Barrier of Obese TNBC Treatments through Suppressing the Interplay between Glycosylation and Ubiquitylation of YAP
Abstract
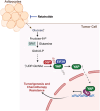
Triple-negative breast cancer (TNBC) in obese patients remains challenging. Recent studies have linked obesity to an increased risk of TNBC and malignancies. Through multiomic analysis and experimental validation, a dysfunctional Eukaryotic Translation Initiation Factor 3 Subunit H (EIF3H)/Yes-associated protein (YAP) proteolytic axis is identified as a pivotal junction mediating the interplay between cancer-associated adipocytes and the response to anti-cancer drugs in TNBC. Mechanistically, cancer-associated adipocytes drive metabolic reprogramming resulting in an upregulated hexosamine biosynthetic pathway (HBP). This aberrant upregulation of HBP promotes YAP O-GlcNAcylation and the subsequent recruitment of EIF3H deubiquitinase, which stabilizes YAP, thus promoting tumor growth and chemotherapy resistance. It is found that Retatrutide, an anti-obesity agent, inhibits HBP and YAP O-GlcNAcylation leading to increased YAP degradation through the deprivation of EIF3H-mediated deubiquitylation of YAP. In preclinical models of obese TNBC, Retatrutide downregulates HBP, decreases YAP protein levels, and consequently decreases tumor size and enhances chemotherapy efficacy. This effect is particularly pronounced in obese mice bearing TNBC tumors. Overall, these findings reveal a critical interplay between adipocyte-mediated metabolic reprogramming and EIF3H-mediated YAP proteolytic control, offering promising therapeutic strategies to mitigate the adverse effects of obesity on TNBC progression.
Keywords: OGT; YAP; adipocyte; deubiquitinase; glycosylation; tumorigenesis.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Dent R., Trudeau M., Pritchard K. I., Hanna W. M., Kahn H. K., Sawka C. A., Lickley L. A., Rawlinson E., Sun P., Narod S. A., Clin. Cancer Res. 2007, 13, 4429. - PubMed
-
- Agarwal G., Nanda G., Lal P., Mishra A., Agarwal A., Agrawal V., Krishnani N., World J. Surg. 2016, 40, 1362. - PubMed
-
- Calle E. E., Rodriguez C., Walker‐Thurmond K., Thun M. J., N. Engl. J. Med. 2003, 348, 1625. - PubMed
-
- Calle E. E., Kaaks R., Nat. Rev. Cancer 2004, 4, 579. - PubMed
-
- Renehan A. G., Tyson M., Egger M., Heller R. F., Zwahlen M., Lancet 2008, 371, 569. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
