Ectopic overexpression of Plasmodium falciparum DNA-/RNA-binding Alba proteins misregulates virulence gene homeostasis during asexual blood development
- PMID: 39868986
- PMCID: PMC11878077
- DOI: 10.1128/spectrum.00885-24
Ectopic overexpression of Plasmodium falciparum DNA-/RNA-binding Alba proteins misregulates virulence gene homeostasis during asexual blood development
Abstract
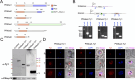
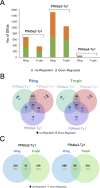
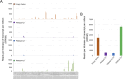
Alba domain-containing proteins are ubiquitously found in archaea and eukaryotes. By binding to either DNA, RNA, or DNA:RNA hybrids, these proteins function in genome stabilization, chromatin organization, gene regulation, and/or translational modulation. In the malaria parasite Plasmodium falciparum, six Alba domain proteins PfAlba1-6 have been described, of which PfAlba1 has emerged as a "master regulator" of translation during parasite intra-erythrocytic development (IED). Given that a tight control of gene expression is especially important during IED, when malaria pathogenesis manifests, in this study, we focus on three other P. falciparum Albas, PfAlba2-4. Because genetic manipulation of the genomic loci of PfAlba2-4 was unsuccessful, we overexpressed each of these proteins from an episome under a strong constitutive promoter. We observed that PfAlba2 or PfAlba3 overexpression strongly reduced parasite growth and impacted IED stage transitions. In contrast, elevated levels of PfAlba4 were well-tolerated by the parasite. In keeping with this, differential gene expression analysis using RNA-seq of PfAlba2 or PfAlba3 overexpressing strains revealed a significant misregulation of mRNAs encoding virulence factors, such as those related to erythrocyte invasion; a general repression of var gene expression was also apparent. PfAlba4 overexpression, on the other hand, did not significantly perturb the steady-state transcriptome of IED stages and appeared to enhance var mRNA levels. Moreover, distinct sets of genes were targeted by each PfAlba for regulation. Taken together, this study highlights the nonredundant roles of PfAlba proteins in the P. falciparum IED, emphasizing their importance in subtelomeric chromatin biology and RNA regulation.IMPORTANCEThe malaria parasite Plasmodium falciparum tightly controls the expression of its genes at the epigenetic, transcriptional, post-transcriptional, and translational levels to synthesize essential proteins, including virulence factors, in a timely and spatially coordinated manner. A family of six proteins implicated in this process is called PfAlba, characterized by the presence of the DNA-, RNA- or DNA:RNA hybrid-binding Alba domain. To better understand the cellular pathways regulated by this protein family, we overexpressed three PfAlbas during P. falciparum intra-erythrocytic growth and found that high levels of PfAlba2 and PfAlba3 were detrimental to parasite development. This was accompanied by significant changes in the parasite's transcriptome, either with regards to mRNA steady-state levels or expression timing. PfAlba4 overexpression, on the other hand, was well-tolerated by the parasite. Overall, our results delineate specific pathways targeted by individual PfAlbas for regulation and link PfAlba2/PfAlba3 to mutually exclusive expression of the virulence-promoting surface antigen PfEMP1.
Keywords: Alba domain; DNA-binding proteins; Plasmodium falciparum; RNA-binding proteins; RNA-seq; gene regulation; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
The PfAlba1 RNA-binding protein is an important regulator of translational timing in Plasmodium falciparum blood stages.Genome Biol. 2015 Sep 28;16:212. doi: 10.1186/s13059-015-0771-5. Genome Biol. 2015. PMID: 26415947 Free PMC article.
-
The Plasmodium falciparum histone methyltransferase PfSET10 is dispensable for the regulation of antigenic variation and gene expression in blood-stage parasites.mSphere. 2024 Nov 21;9(11):e0054624. doi: 10.1128/msphere.00546-24. Epub 2024 Oct 24. mSphere. 2024. PMID: 39445826 Free PMC article.
-
The RNA structurome in the asexual blood stages of malaria pathogen plasmodium falciparum.RNA Biol. 2021 Dec;18(12):2480-2497. doi: 10.1080/15476286.2021.1926747. Epub 2021 Jun 23. RNA Biol. 2021. PMID: 33960872 Free PMC article.
-
The role of PfEMP1 adhesion domain classification in Plasmodium falciparum pathogenesis research.Mol Biochem Parasitol. 2014 Jul;195(2):82-7. doi: 10.1016/j.molbiopara.2014.07.006. Epub 2014 Jul 23. Mol Biochem Parasitol. 2014. PMID: 25064606 Free PMC article. Review.
-
Plasmodium falciparum: epigenetic control of var gene regulation and disease.Subcell Biochem. 2013;61:659-82. doi: 10.1007/978-94-007-4525-4_28. Subcell Biochem. 2013. PMID: 23150271 Review.
Cited by
-
Evolution of sequence, structural and functional diversity of the ubiquitous DNA/RNA-binding Alba domain.Sci Rep. 2024 Dec 5;14(1):30363. doi: 10.1038/s41598-024-79937-4. Sci Rep. 2024. PMID: 39638848 Free PMC article.
References
-
- Le Roch KG, Johnson JR, Florens L, Zhou Y, Santrosyan A, Grainger M, Yan SF, Williamson KC, Holder AA, Carucci DJ, Yates JR III, Winzeler EA. 2004. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res 14:2308–2318. doi:10.1101/gr.2523904 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

