Characterisation of RNA guanine-7 methyltransferase (RNMT) using a small molecule approach
- PMID: 39869500
- PMCID: PMC12133303
- DOI: 10.1042/BCJ20240608
Characterisation of RNA guanine-7 methyltransferase (RNMT) using a small molecule approach
Abstract
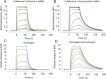
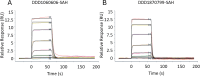
The maturation of the RNA cap involving guanosine N-7 methylation, catalyzsed by the HsRNMT (RNA guanine-7 methyltransferase (HsRNMT)-RAM (RNA guanine-N7 methyltransferase activating subunit (RAM) complex, is currently under investigation as a novel strategy to combat PIK3CA -mutant breast cancer. However, the development of effective drugs is hindered by a limited understanding of the enzyme's mechanism and a lack of small molecule inhibitors. Following the elucidation of the HsRNMT-RAM molecular mechanism, we report the biophysical characterizsation of two small molecule hits. Biophysics, biochemistry and structural biology confirm that both compounds bind competitively with cap and bind effectively to HsRNMT-RAM in the presence of the co-product SAH, with a binding affinity (KD) of approximately 1 μM. This stabilisation of the enzyme--product complex results in uncompetitive inhibition. Finally, we describe the properties of the cap pocket and provided suggestions for further development of the tool compounds.
Keywords: RNMT; Sinefungin; breast cancer; methyl transferase.
© 2025 The Author(s).
Conflict of interest statement
The Authors declare that there are no competing interests associated with this manuscript.
Figures





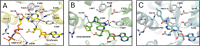



Similar articles
-
Mechanism of allosteric activation of human mRNA cap methyltransferase (RNMT) by RAM: insights from accelerated molecular dynamics simulations.Nucleic Acids Res. 2019 Sep 19;47(16):8675-8692. doi: 10.1093/nar/gkz613. Nucleic Acids Res. 2019. PMID: 31329932 Free PMC article.
-
Molecular basis of RNA guanine-7 methyltransferase (RNMT) activation by RAM.Nucleic Acids Res. 2016 Dec 1;44(21):10423-10436. doi: 10.1093/nar/gkw637. Epub 2016 Jul 15. Nucleic Acids Res. 2016. PMID: 27422871 Free PMC article.
-
RAM function is dependent on Kapβ2-mediated nuclear entry.Biochem J. 2014 Feb 1;457(3):473-84. doi: 10.1042/BJ20131359. Biochem J. 2014. PMID: 24200467 Free PMC article.
-
The RNA cap methyltransferases RNMT and CMTR1 co-ordinate gene expression during neural differentiation.Biochem Soc Trans. 2023 Jun 28;51(3):1131-1141. doi: 10.1042/BST20221154. Biochem Soc Trans. 2023. PMID: 37145036 Free PMC article. Review.
-
H3K36 methyltransferases as cancer drug targets: rationale and perspectives for inhibitor development.Future Med Chem. 2016 Sep;8(13):1589-607. doi: 10.4155/fmc-2016-0071. Epub 2016 Aug 22. Future Med Chem. 2016. PMID: 27548565 Free PMC article. Review.
Cited by
-
Uncovering the Epitranscriptome: A Review on mRNA Modifications and Emerging Frontiers.Genes (Basel). 2025 Aug 12;16(8):951. doi: 10.3390/genes16080951. Genes (Basel). 2025. PMID: 40870000 Free PMC article. Review.
-
Structural basis for sensitivity and acquired resistance of fungal cap guanine-N7 methyltransferases to the antifungal antibiotic sinefungin.Nucleic Acids Res. 2025 Jul 19;53(14):gkaf538. doi: 10.1093/nar/gkaf538. Nucleic Acids Res. 2025. PMID: 40682821 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

