Splicing accuracy varies across human introns, tissues, age and disease
- PMID: 39870615
- PMCID: PMC11772838
- DOI: 10.1038/s41467-024-55607-x
Splicing accuracy varies across human introns, tissues, age and disease
Abstract
Alternative splicing impacts most multi-exonic human genes. Inaccuracies during this process may have an important role in ageing and disease. Here, we investigate splicing accuracy using RNA-sequencing data from >14k control samples and 40 human body sites, focusing on split reads partially mapping to known transcripts in annotation. We show that splicing inaccuracies occur at different rates across introns and tissues and are affected by the abundance of core components of the spliceosome assembly and its regulators. We find that age is positively correlated with a global decline in splicing fidelity, mostly affecting genes implicated in neurodegenerative diseases. We find support for the latter by observing a genome-wide increase in splicing inaccuracies in samples affected with Alzheimer's disease as compared to neurologically normal individuals. In this work, we provide an in-depth characterisation of splicing accuracy, with implications for our understanding of the role of inaccuracies in ageing and neurodegenerative disorders.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: S.G. is a current employee of Verge Genomics. All work performed for this publication was performed in his own time, and not as a part of his duties as an employee. R.H.R and D.Z are current employees of CoSyne Therapeutics. All work performed for this publication was performed in their own time, and not as a part of their duties as employees. The remaining authors declare no competing interests.
Figures

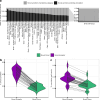
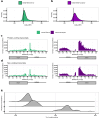

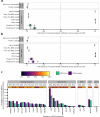

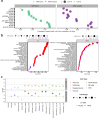

Update of
-
Splicing accuracy varies across human introns, tissues and age.bioRxiv [Preprint]. 2023 Mar 30:2023.03.29.534370. doi: 10.1101/2023.03.29.534370. bioRxiv. 2023. Update in: Nat Commun. 2025 Jan 27;16(1):1068. doi: 10.1038/s41467-024-55607-x. PMID: 37034741 Free PMC article. Updated. Preprint.
References
-
- Shi, Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol.18, 655–670 (2017). - PubMed
-
- Chow, L. T., Gelinas, R. E., Broker, T. R. & Roberts, R. J. An amazing sequence arrangement at the 5’ ends of adenovirus 2 messenger RNA. Cell12, 1–8 (1977). - PubMed
MeSH terms
Grants and funding
- ASAP-000486/Michael J. Fox Foundation for Parkinson's Research (Michael J. Fox Foundation)
- ASAP-000509/Michael J. Fox Foundation for Parkinson's Research (Michael J. Fox Foundation)
- A2021009F/BrightFocus Foundation (BrightFocus)
- R01MH123567/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- MR/N008324/1/RCUK | Medical Research Council (MRC)

