A compendium of Amplification-Related Gain Of Sensitivity genes in human cancer
- PMID: 39870664
- PMCID: PMC11772776
- DOI: 10.1038/s41467-025-56301-2
A compendium of Amplification-Related Gain Of Sensitivity genes in human cancer
Abstract
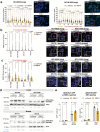
While the effect of amplification-induced oncogene expression in cancer is known, the impact of copy-number gains on "bystander" genes is less understood. We create a comprehensive map of dosage compensation in cancer by integrating expression and copy number profiles from over 8000 tumors in The Cancer Genome Atlas and cell lines from the Cancer Cell Line Encyclopedia. Additionally, we analyze 17 cancer open reading frame screens to identify genes toxic to cancer cells when overexpressed. Combining these approaches, we propose a class of 'Amplification-Related Gain Of Sensitivity' (ARGOS) genes located in commonly amplified regions, yet expressed at lower levels than expected by their copy number, and toxic when overexpressed. We validate RBM14 as an ARGOS gene in lung and breast cancer cells, and suggest a toxicity mechanism involving altered DNA damage response and STING signaling. We additionally observe increased patient survival in a radiation-treated cancer cohort with RBM14 amplification.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: K.S. is on the SAB and has stock options with Auron Therapeutics. K.S. receives grant funding from Novartis and KronosBio on topics unrelated to this manuscript. W.C.H. is a consultant for Thermo Fischer Scientific, Solasta Ventures, MPM capital, KSQ Therapeutics, Tyra Biosciences, Frontier Medicines, Jubilant Therapeutics, RAPPTA Therapeutics, Serinus Biosciences, Hexagon Biosciences, Kestral Therapeutics, Function Oncology, and Calyx. R.B. consults for Scorpion Therapeutics and receives grant funding from Novartis. U.B.D. consults for Accenet Therapeutics and received funding from Novocure. F.F. is co-founder and Chief Scientific Officer of iPsomics. The work in this study is unrelated to this role. The remaining authors declare no competing interests.
Figures






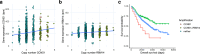
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

