Next generation sequencing of multiple SARS-CoV-2 infections in the Omicron Era
- PMID: 39870695
- PMCID: PMC11772649
- DOI: 10.1038/s41598-024-84952-6
Next generation sequencing of multiple SARS-CoV-2 infections in the Omicron Era
Abstract
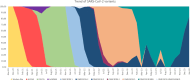
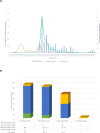
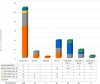
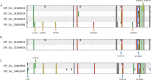
Since the emergence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the need for an effective vaccine has appeared crucial for stimulating immune system responses to produce humoral/cellular immunity and activate immunological memory. It has been demonstrated that SARS-CoV-2 variants escape neutralizing immunity elicited by previous infection and/or vaccination, leading to new infection waves and cases of reinfection. The study aims to gain into cases of reinfections, particularly infections and/or vaccination-induced protection. We conducted a retrospective descriptive study using data collected during the SARS-CoV-2 pandemic. This analysis involved Reverse Transcriptase Quantitative Polymerase Chain Reaction (RT-qPCR) and Next Generation Sequencing (NGS). RT-qPCR was performed on 416,466 naso-oropharyngeal swabs, with 10,380 samples further analyzed using NGS technology. RT-qPCR identified 350 cases of reinfection, of which 228 were subjected to detailed analysis via NGS. Our findings revealed two interesting cases involving pediatric patients who were not vaccinated. Positive results were observed in these cases within a short interval (< 60 days) and the "nature" of the infection, whether attributable to Reinfection or Viral Persistence, was investigated. Specifically, we discuss a case involving an unvaccinated 18-month-old child, which may represent one of the earliest instances of BA.5/BA.5 reinfection identified worldwide.
Keywords: NGS; Omicron; Reactivation; Reinfection; SARS-CoV-2; Vaccine.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Effect of wild-type vaccine doses on BA.5 hybrid immunity, disease severity, and XBB reinfection risk.J Virol. 2024 Dec 17;98(12):e0128524. doi: 10.1128/jvi.01285-24. Epub 2024 Nov 5. J Virol. 2024. PMID: 39499071 Free PMC article.
-
Dynamic monitoring of antibody titers in people living with HIV during Omicron epidemic: comparison between unvaccinated and vaccinated individuals.BMC Infect Dis. 2025 Jul 30;25(1):962. doi: 10.1186/s12879-025-11387-3. BMC Infect Dis. 2025. PMID: 40739195 Free PMC article.
-
Saline gargle collection method is comparable to nasopharyngeal/oropharyngeal swabbing for the molecular detection and sequencing of SARS-CoV-2 in Botswana.Microbiol Spectr. 2025 Jul;13(7):e0202324. doi: 10.1128/spectrum.02023-24. Epub 2025 May 22. Microbiol Spectr. 2025. PMID: 40401962 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
SARS-CoV-2-neutralising monoclonal antibodies to prevent COVID-19.Cochrane Database Syst Rev. 2022 Jun 17;6(6):CD014945. doi: 10.1002/14651858.CD014945.pub2. Cochrane Database Syst Rev. 2022. PMID: 35713300 Free PMC article.
References
-
- European Centre for Disease Prevention and Control. https://www.ecdc.europa.eu/en/publications-data/reinfection-SARS-CoV-2-i... (2021).
-
- Markov, P. V. et al. The evolution of SARS-CoV-2. Nat. Rev. Microbiol.21, 361–379. 10.1038/s41579-023-00878-2 (2023). - PubMed
-
- World Health Organization. https://www.who.int/activities/tracking-SARS-CoV-2-variants (2024).
-
- World Health Organization. http://www.who.int/news/item/16-03-2023-statement-on-the-update-of-who-s... (2023).
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

