Preclinical use of a clinically-relevant scAAV9/SUMF1 vector for the treatment of multiple sulfatase deficiency
- PMID: 39870870
- PMCID: PMC11772666
- DOI: 10.1038/s43856-025-00734-9
Preclinical use of a clinically-relevant scAAV9/SUMF1 vector for the treatment of multiple sulfatase deficiency
Abstract
Background: Multiple Sulfatase Deficiency (MSD) is a rare inherited lysosomal storage disorder characterized by loss of function mutations in the SUMF1 gene that manifests as a severe pediatric neurological disease. There are no available targeted therapies for MSD.
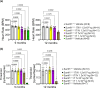
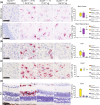
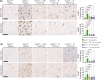
Methods: We engineered a viral vector (AAV9/SUMF1) to deliver working copies of the SUMF1 gene and tested the vector in Sumf1 knock out mice that generally display a median lifespan of 10 days. Mice were injected as pre-symptomatic neonates via intracerebroventricular administration, or as post-symptomatic juveniles via intrathecal alone or combination intrathecal and intravenous delivery. Cohorts were assessed for survival, behavioral outcomes, and post-mortem for sulfatase activity.
Results: We show that treatment of neonates extends survival up to 1-year post-injection. Importantly, delivery of SUMF1 through cerebral spinal fluid at 7 days of age alleviates MSD symptoms. The treated mice show wide distribution of the SUMF1 gene, no signs of toxicity or neuropathy, improved vision and cardiac function, and no behavioral deficits. One-year post treatment, tissues show increased sulfatase activity, indicating functional SUMF1. Further, a GLP toxicology study conducted in rats demonstrates favorable overall safety of this approach.
Conclusions: These preclinical studies highlight the potential of our AAV9/SUMF1 vector, the design of which is directly translatable for clinical use, as a gene replacement therapy for MSD patients.
Plain language summary
Multiple Sulfatase Deficiency (MSD) is a rare genetic disorder caused by mutations in the SUMF1 gene, leading to severe problems with the brain and other organs in children. Currently, there is no available treatment. Here we tested an approach called gene therapy in which the deficient Sumf1 gene is replaced with the normal gene in a mouse model of MSD. Mice treated shortly after birth showed no major problems in brain and other organs and lived up to a year, while those treated after symptoms appeared also lived longer, with over half surviving beyond 15 months. This study suggests that this gene therapy approach could be a promising and safe treatment for MSD.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: R.M.B and S.J.G. received royalty income from Neurogene Inc. and Taysha Gene Therapies (World patent: WO2020223215A1, “Optimized SUMF1 Genes and Expression Cassettes and Their Use”). The other authors declare no competing interests.
Figures




References
-
- Cosma, M. P. et al. The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell113, 445–456 (2003). - PubMed
-
- Dierks, T. et al. Multiple sulfatase deficiency is caused by mutations in the gene encoding the human Cα-formylglycine generating enzyme. Cell113, 435–444 (2003). - PubMed
-
- Dierks, T. et al. Molecular basis for multiple sulfatase deficiency and mechanism for formylglycine generation of the human formylglycine-generating enzyme. Cell121, 541–552 (2005). - PubMed
-
- Cappuccio, G., Alagia, M. & Brunetti-Pierri, N. A systematic cross-sectional survey of multiple sulfatase deficiency. Mol. Genet. Metab.130, 283–288 (2020). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

