Asymmetries in the Architecture of ON and OFF Arbors in ON-OFF Direction-Selective Ganglion Cells
- PMID: 39871013
- PMCID: PMC7617701
- DOI: 10.1002/cne.70023
Asymmetries in the Architecture of ON and OFF Arbors in ON-OFF Direction-Selective Ganglion Cells
Abstract
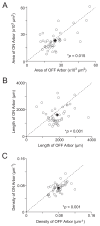
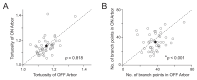
Direction selectivity is a fundamental feature in the visual system. In the retina, direction selectivity is independently computed by ON and OFF circuits. However, the advantages of extracting directional information from these two independent circuits are unclear. To gain insights, we examined the ON-OFF direction-selective ganglion cells (DSGCs), which recombine signals from both circuits. Specifically, we investigated the dendritic architecture of these neurons with the premise that asymmetries in architecture will provide insights into function. Scrutinizing the dendrites of dye-filled ON-OFF DSGCs reveals that the OFF arbors of these neurons are substantially denser. The increase in density can be primarily attributed to the higher branching seen in OFF arbors. Further, analysis of ON-OFF DSGCs in a previously published serial block-face electron microscopy dataset revealed that the denser OFF arbors packed more bipolar synapses per unit dendritic length. These asymmetries in the dendritic architecture suggest that the ON-OFF DSGC preferentially magnifies the synaptic drive of the OFF pathway, potentially allowing it to encode information distinct from the ON pathway.
Keywords: ON–OFF; dendritic architecture; dendritic density; direction selectivity; morphology; retinal ganglion cell; synaptic inputs.
© 2025 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Differential Expression Analysis Identifies Candidate Synaptogenic Molecules for Wiring Direction-Selective Circuits in the Retina.J Neurosci. 2024 May 1;44(18):e1461232024. doi: 10.1523/JNEUROSCI.1461-23.2024. J Neurosci. 2024. PMID: 38514178 Free PMC article.
-
Divergent mechanisms of neural adaptation and instability in the mammalian retina.Curr Biol. 2025 Jul 21;35(14):3381-3395.e4. doi: 10.1016/j.cub.2025.06.015. Epub 2025 Jun 27. Curr Biol. 2025. PMID: 40580962
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of topotecan for ovarian cancer.Health Technol Assess. 2001;5(28):1-110. doi: 10.3310/hta5280. Health Technol Assess. 2001. PMID: 11701100
-
Factors that impact on the use of mechanical ventilation weaning protocols in critically ill adults and children: a qualitative evidence-synthesis.Cochrane Database Syst Rev. 2016 Oct 4;10(10):CD011812. doi: 10.1002/14651858.CD011812.pub2. Cochrane Database Syst Rev. 2016. PMID: 27699783 Free PMC article.
-
Education support services for improving school engagement and academic performance of children and adolescents with a chronic health condition.Cochrane Database Syst Rev. 2023 Feb 8;2(2):CD011538. doi: 10.1002/14651858.CD011538.pub2. Cochrane Database Syst Rev. 2023. PMID: 36752365 Free PMC article.
References
-
- Amthor FR, Oyster CW, Takahashi ES. Morphology of ON-OFF Direction-Selective Ganglion Cells in the Rabbit Retina. Brain Research. 1984;298:187–190. - PubMed
-
- Amthor FR, Takahashi ES, Oyster CW. Morphologies of Rabbit Retinal Ganglion Cells With Complex Receptive Fields. Journal of Comparative Neurology. 1989;280:97–121. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

