MoCloro: an extension of the Chlamydomonas reinhardtii modular cloning toolkit for microalgal chloroplast engineering
- PMID: 39871102
- PMCID: PMC11772913
- DOI: 10.1111/ppl.70088
MoCloro: an extension of the Chlamydomonas reinhardtii modular cloning toolkit for microalgal chloroplast engineering
Abstract
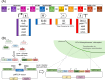
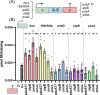
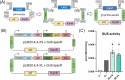
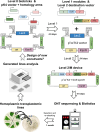
Photosynthetic microalgae are promising green cell factories for the sustainable production of high-value chemicals and biopharmaceuticals. The chloroplast organelle is being developed as a chassis for synthetic biology as it contains its own genome (the plastome) and some interesting advantages, such as high recombinant protein titers and a diverse and dynamic metabolism. However, chloroplast engineering is currently hampered by the lack of standardized cloning tools and Design-Build-Test-Learn workflows to ease genomic and metabolic engineering. The MoClo (Modular Cloning) toolkit based on Golden Gate assembly was recently developed in the model eukaryotic green microalgae Chlamydomonas reinhardtii to facilitate nuclear transformation and engineering. Here, we present MoCloro as an extension of the MoClo that allows chloroplast genome engineering. Briefly, a Golden Gate-compatible chloroplast transformation vector (pWF.K.4) was constructed, which contains homologous arms for integration at the petA site in the plastome. A collection of standardized parts (promoters, terminators, reporter and selection marker genes) was created following the MoClo syntax to enable easy combinatorial assembly of multi-cassettes in the destination pWF.K.4 vector. The functionality of the biobricks was assayed by constructing and assessing the expression of several multigenic constructs. Finally, a generic vector pK4 was constructed for easy Golden Gate cloning of 5' and 3' homologous arms, allowing targeting at alternative plastome integration sites. This work represents a significant advancement in technology aimed at facilitating more efficient and rapid chloroplast transformation and engineering of green microalgae.
© 2025 The Author(s). Physiologia Plantarum published by John Wiley & Sons Ltd on behalf of Scandinavian Plant Physiology Society.
Figures

Similar articles
-
Multigenic engineering of the chloroplast genome in the green alga Chlamydomonas reinhardtii.Microbiology (Reading). 2020 Jun;166(6):510-515. doi: 10.1099/mic.0.000910. Microbiology (Reading). 2020. PMID: 32250732 Free PMC article.
-
Birth of a Photosynthetic Chassis: A MoClo Toolkit Enabling Synthetic Biology in the Microalga Chlamydomonas reinhardtii.ACS Synth Biol. 2018 Sep 21;7(9):2074-2086. doi: 10.1021/acssynbio.8b00251. Epub 2018 Sep 5. ACS Synth Biol. 2018. PMID: 30165733
-
Rational Design and Screening of Synthetic Promoters in Chlamydomonas reinhardtii.Methods Mol Biol. 2024;2844:69-83. doi: 10.1007/978-1-0716-4063-0_4. Methods Mol Biol. 2024. PMID: 39068332
-
Green biologics: The algal chloroplast as a platform for making biopharmaceuticals.Bioengineered. 2018 Jan 1;9(1):48-54. doi: 10.1080/21655979.2017.1377867. Epub 2017 Sep 29. Bioengineered. 2018. PMID: 28892417 Free PMC article. Review.
-
Manipulation of the microalgal chloroplast by genetic engineering for biotechnological utilization as a green biofactory.World J Microbiol Biotechnol. 2018 Nov 26;34(12):183. doi: 10.1007/s11274-018-2567-8. World J Microbiol Biotechnol. 2018. PMID: 30478596 Review.
Cited by
-
A game of tag: A review of protein tags for the successful detection, purification and fluorescence labelling of proteins expressed in microalgae.Plant J. 2025 Jun;122(6):e70272. doi: 10.1111/tpj.70272. Plant J. 2025. PMID: 40537033 Free PMC article. Review.
References
-
- Dauvillee, D. et al. (2004) Minimal extent of sequence homology required for homologous recombination at the psbA locus in Chlamydomonas reinhardtii chloroplasts using PCR‐generated DNA fragments, Photosynthesis Research. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

