Prevalence and dynamics of antimicrobial resistance in pioneer and developing Arctic soils
- PMID: 39871128
- PMCID: PMC11771051
- DOI: 10.1186/s12866-025-03745-7
Prevalence and dynamics of antimicrobial resistance in pioneer and developing Arctic soils
Abstract
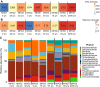
Antimicrobial resistance (AMR) in soil is an ancient phenomenon with widespread spatial presence in terrestrial ecosystems. However, the natural processes shaping the temporal dissemination of AMR in soils are not well understood. We aimed to determine whether, how, and why AMR varies with soil age in recently deglaciated pioneer and developing Arctic soils using a space-for-time approach. Specifically, we assess how the magnitude and spread of AMR changes with soil development stages, including antibiotic resistance genes (ARGs), mobile genetic elements (MGEs), and antibiotic-resistant bacteria (ARB). We showed that ARGs, MGEs, and ARB are present, and exhibit a non-uniform distribution in the developing soils. Their abundance generally increases with soil age but at different rates overall and across different glacier forefields. Our analyses suggest a strong positive relationship between soil age and ARGs and ARB, which we attribute to increased competition between microbes in older soils. We also observed a strong negative relationship between soil age and ARG diversity mediated by soil organic matter - suggesting facilitation due to the alleviation of nutrient limitation. These contrasting results suggest that both competition and facilitation can regulate AMR spread through time in the Arctic, but competition might be the stronger determinant of AMR spread.
Keywords: Antimicrobial resistance; Arctic soils; Metagenomics; Microorganisms; Soil development.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Clinical trial number: not applicable. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Field-based evidence for enrichment of antibiotic resistance genes and mobile genetic elements in manure-amended vegetable soils.Sci Total Environ. 2019 Mar 1;654:906-913. doi: 10.1016/j.scitotenv.2018.10.446. Epub 2018 Nov 3. Sci Total Environ. 2019. PMID: 30453260
-
Investigation of high-risk antibiotic resistance bacteria and their associated antibiotic resistance genes in different agricultural soils with biogas slurry from China.J Hazard Mater. 2024 Aug 5;474:134775. doi: 10.1016/j.jhazmat.2024.134775. Epub 2024 May 31. J Hazard Mater. 2024. PMID: 38824772
-
Understanding drivers of antibiotic resistance genes in High Arctic soil ecosystems.Environ Int. 2019 Apr;125:497-504. doi: 10.1016/j.envint.2019.01.034. Epub 2019 Jan 28. Environ Int. 2019. PMID: 30700387
-
Sources of Antibiotic Resistant Bacteria (ARB) and Antibiotic Resistance Genes (ARGs) in the Soil: A Review of the Spreading Mechanism and Human Health Risks.Rev Environ Contam Toxicol. 2021;256:121-153. doi: 10.1007/398_2020_60. Rev Environ Contam Toxicol. 2021. PMID: 33948742 Review.
-
Micro-interfacial behavior of antibiotic-resistant bacteria and antibiotic resistance genes in the soil environment: A review.Environ Int. 2024 Sep;191:108972. doi: 10.1016/j.envint.2024.108972. Epub 2024 Aug 21. Environ Int. 2024. PMID: 39180776 Review.
References
-
- Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010;8:251–9. - PubMed
-
- D’Costa VM, King CE, Kalan L, Morar M, Sung WWL, Schwarz C, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–61. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

