The quest for environmental analytical microbiology: absolute quantitative microbiome using cellular internal standards
- PMID: 39871306
- PMCID: PMC11773863
- DOI: 10.1186/s40168-024-02009-2
The quest for environmental analytical microbiology: absolute quantitative microbiome using cellular internal standards
Abstract
Background: High-throughput sequencing has revolutionized environmental microbiome research, providing both quantitative and qualitative insights into nucleic acid targets in the environment. The resulting microbial composition (community structure) data are essential for environmental analytical microbiology, enabling characterization of community dynamics and assessing microbial pollutants for the development of intervention strategies. However, the relative abundances derived from sequencing impede comparisons across samples and studies.
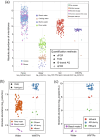
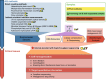
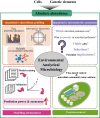
Results: This review systematically summarizes various absolute quantification (AQ) methods and their applications to obtain the absolute abundance of microbial cells and genetic elements. By critically comparing the strengths and limitations of AQ methods, we advocate the use of cellular internal standard-based high-throughput sequencing as an appropriate AQ approach for studying environmental microbiome originated from samples of complex matrices and high heterogeneity. To minimize ambiguity and facilitate cross-study comparisons, we outline essential reporting elements for technical considerations, and provide a checklist as a reference for environmental microbiome research.
Conclusions: In summary, we propose absolute microbiome quantification using cellular internal standards for environmental analytical microbiology, and we anticipate that this approach will greatly benefit future studies. Video Abstract.
Keywords: Absolute quantification; Cellular internal standard; Environmental analytical microbiology; Microbiome; Standardization.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The manuscript does not report data collected from humans and animals. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Towards Quantitative Microbiome Community Profiling Using Internal Standards.Appl Environ Microbiol. 2019 Feb 20;85(5):e02634-18. doi: 10.1128/AEM.02634-18. Print 2019 Mar 1. Appl Environ Microbiol. 2019. PMID: 30552195 Free PMC article.
-
Gradient Internal Standard Method for Absolute Quantification of Microbial Amplicon Sequencing Data.mSystems. 2021 Jan 12;6(1):e00964-20. doi: 10.1128/mSystems.00964-20. mSystems. 2021. PMID: 33436513 Free PMC article.
-
Synthetic spike-in standards for high-throughput 16S rRNA gene amplicon sequencing.Nucleic Acids Res. 2017 Feb 28;45(4):e23. doi: 10.1093/nar/gkw984. Nucleic Acids Res. 2017. PMID: 27980100 Free PMC article.
-
The quest for absolute abundance: The use of internal standards for DNA-based community ecology.Mol Ecol Resour. 2021 Jan;21(1):30-43. doi: 10.1111/1755-0998.13247. Epub 2020 Sep 17. Mol Ecol Resour. 2021. PMID: 32889760 Review.
-
What Is Metagenomics Teaching Us, and What Is Missed?Annu Rev Microbiol. 2020 Sep 8;74:117-135. doi: 10.1146/annurev-micro-012520-072314. Epub 2020 Jun 30. Annu Rev Microbiol. 2020. PMID: 32603623 Review.
References
-
- Yang Y, Che Y, Liu L, Wang C, Yin X, Deng Y, et al. Rapid absolute quantification of pathogens and ARGs by nanopore sequencing. Sci Total Environ. 2021;809:152190. - PubMed
-
- Yin X, Yang Y, Deng Y, Huang Y, Li L, Chan LY, et al. An assessment of resistome and mobilome in wastewater treatment plants through temporal and spatial metagenomic analysis. Water Res. 2022;209:117885. - PubMed
-
- Evans PN, Boyd JA, Leu AO, Woodcroft BJ, Parks DH, Hugenholtz P, et al. An evolving view of methane metabolism in the Archaea. Nat Rev Microbiol. 2019;17(4):219–32. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

