The fungal microbiota modulate neonatal oxygen-induced lung injury
- PMID: 39871397
- PMCID: PMC11773857
- DOI: 10.1186/s40168-025-02032-x
The fungal microbiota modulate neonatal oxygen-induced lung injury
Abstract
Background: The immature lungs of very preterm infants are exposed to supraphysiologic oxygen, contributing to bronchopulmonary dysplasia (BPD), a chronic lung disease that is the most common morbidity of prematurity. While the microbiota significantly influences neonatal health, the relationship between the intestinal microbiome, particularly micro-eukaryotic members such as fungi and yeast, and lung injury severity in newborns remains unknown.
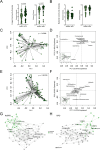
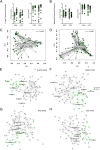
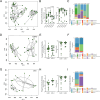
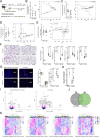
Results: Here, we show that the fungal microbiota modulates hyperoxia-induced lung injury severity in very low birth weight premature infants and preclinical pseudohumanized and altered fungal colonization mouse models. Instead of fungal communities dominated by Candida and Saccharomyces, the first stool microbiomes of infants who developed BPD had less interconnected community architectures with a greater diversity of rarer fungi. After using a pseudohumanized model to show that transfer to the neonatal microbiome from infants with BPD increased the severity of lung injury, we used gain and loss of function approaches to demonstrate that modulating the extent of initial neonatal fungal colonization affected the extent of BPD-like lung injury in mice. We also identified alterations in the murine intestinal microbiome and transcriptome associated with augmented lung injury.
Conclusions: These findings demonstrate that features of the initial intestinal fungal microbiome are associated with the later development of BPD in premature neonates and exert a microbiome-driven effect that is transferable and modifiable in murine models, which suggests both causality and a potential therapeutic strategy. Video Abstract.
Keywords: Chronic lung injury; Fungal microbiome; Gut microbiome; Multikingdom microbiome; Mycobiome; Preterm infant; Very low birthweight; Yeast.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The human study protocol was approved by the Institutional Review Board of The University of Tennessee under protocol 17–05311-XP and conducted in accordance with the Declaration of Helsinki. The human cohort for FMT samples was approved under protocol IRB-300003994 by the IRB at the University of Alabama at Birmingham (UAB, Birmingham, AL, USA). Mouse experiments were performed at UAB under IACUC protocol 22042. Consent for publication: Assent for publication was obtained from the mother during study enrollment. Competing interests: CL is the founder of ResBiotic Nutrition, Inc. and Alveolus Bio, Inc., from which he reports salary and stock options, and TN is now an employee, thereof. KW and NA are scientific advisors to these companies and report consulting fees and stock options from the same. The remaining authors report no disclosures.
Figures

References
-
- Lui K, Lee SK, Kusuda S, Adams M, Vento M, Reichman B, et al. Trends in outcomes for neonates born very preterm and very low birth weight in 11 high-income countries. J Pediatr. 2019;215:32-40.e14. - PubMed
-
- Kinsella JP, Greenough A, Abman SH. Bronchopulmonary dysplasia. Lancet. 2006;367:1421–31. - PubMed
-
- Dang AT, Marsland BJ. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 2019;12:843–50. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

