Single-cell RNA-seq reveals immune cell heterogeneity and increased Th17 cells in human fibrotic skin diseases
- PMID: 39872534
- PMCID: PMC11769821
- DOI: 10.3389/fimmu.2024.1522076
Single-cell RNA-seq reveals immune cell heterogeneity and increased Th17 cells in human fibrotic skin diseases
Abstract
Background: Fibrotic skin disease represents a major global healthcare burden, characterized by fibroblast hyperproliferation and excessive accumulation of extracellular matrix components. The immune cells are postulated to exert a pivotal role in the development of fibrotic skin disease. Single-cell RNA sequencing has been used to explore the composition and functionality of immune cells present in fibrotic skin diseases. However, these studies detected the gene expression of all cells in fibrotic skin diseases and did not enrich immune cells. Thus, the precise immune cell atlas in fibrotic skin diseases remains unknown. In this study, we plan to investigate the intricate cellular landscape of immune cells in keloid, a paradigm of fibrotic skin diseases.
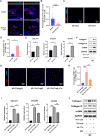
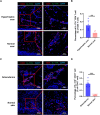
Methods: CD45+ immune cells were enriched by fluorescence-activated cell sorting. Single-cell RNA sequencing was used to analyze the cellular landscape of immune cells in keloid and normal scar tissues. Ki-67 staining, a scratch experiment, real-time PCR, and Western blotting were used to explore the effect of the Th17 cell supernatant on keloid fibroblasts.
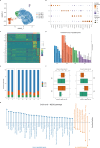
Results: Our findings revealed the intricate cellular landscape of immune cells in fibrotic skin diseases. We found that the percentage of Th17 cells was significantly increased in keloids compared to normal scars. All the subclusters of macrophages and dendritic cells (DCs) showed similar proportions between keloid samples and normal scar samples. However, upregulated genes in keloid M1 macrophages, M2 macrophages, and cDC2 are associated with the MHC class II protein complex assembly and antigen assembly, indicating that macrophages and cDC2 are active in keloids. Functional studies suggested that the supernatant of Th17 cells could promote proliferation, collagen expression, and migration of keloid fibroblasts through interleukin 17A. Importantly, increased Th17 cells are also found in other fibrotic skin diseases, such as hypertrophic scars and scleroderma, suggesting this represents a broad mechanism for skin fibrosis.
Conclusion: In summary, we built a single-cell atlas of fibrotic skin diseases in this study. In addition, we explored the function of Th17 cell-fibroblast interaction in skin fibrosis. These findings will help to understand fibrotic skin disease pathogenesis in depth and identify potential targets for fibrotic skin disease treatment.
Keywords: IL-17; Th17 cell; dendritic cell; fibrotic skin diseases; immune cell; keloid; macrophage.
Copyright © 2025 Deng, Xu, Zhang, Liu, Wang, Chen, Yao, Zhu and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

