Learning From Full Characterization of HIV Proviruses in People Receiving Long-Acting Cabotegravir/Rilpivirine With a History of Replication on the Antiretroviral Classes
- PMID: 39872809
- PMCID: PMC11770275
- DOI: 10.1093/ofid/ofae748
Learning From Full Characterization of HIV Proviruses in People Receiving Long-Acting Cabotegravir/Rilpivirine With a History of Replication on the Antiretroviral Classes
Abstract
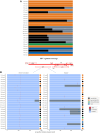
Background: To better understand factors associated with virologic response, we retrospectively characterized the HIV proviruses of 7 people with HIV who received long-acting cabotegravir/rilpivirine (CAB/RPV-LA) and were selected according to the following criteria: virologic control achieved despite a history of viral replication on 1 or both corresponding antiretroviral classes (n = 6) and virologic failure (VF) after CAB/RPV-LA initiation (n = 1).
Methods: Last available blood samples before the initiation of CAB/RPV-LA were analyzed retrospectively. Near full-length HIV DNA genome haplotypes were inferred from Nanopore sequencing by the in vivo Genome Diversity Analyzer to search for archived drug resistance mutations (DRMs) and evaluate the frequency and intactness of proviruses harboring DRMs.
Results: Archived DRMs including G-to-A mutations were found in samples from 3 patients who maintained virologic control. Genomes harboring DRMs were majorly in minority variants (<20%) and were defective in all cases except for 1 participant. In this participant, intact genomes with the H221Y mutation on reverse transcriptase were detected representing 11 copies per 106 peripheral blood mononuclear cells. The other mutations observed in the participants of the study resulted most likely from hypermutations. The patient with VF presented archived mutations, all associated with defects. Other factors could explain this VF.
Conclusions: Our findings highlight the difficulty in interpreting the clinical significance of DRMs when detected in proviral DNA and the need to filter out hypermutated sequences. Detected DRMs could be harbored by defective archived genomes unlikely to contribute to treatment failure.
Keywords: Drug Resistance Mutations; HIV; HIV DNA genotyping; long-acting antiretrovirals; near-full genome sequencing.
© The Author(s) 2024. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest. V. A.-F. has received institutional grants from ViiV Healthcare and honoraria and travel grants from ViiV Healthcare and Gilead Sciences for participation in educational meetings and conferences, all outside the submitted work. L. H. reports nonfinancial support from Gilead, Merck Sharp & Dohme, and ViiV Healthcare; honoraria payments and travel support for advisory board participation from Gilead, Merck Sharp & Dohme, and ViiV Healthcare; and personal consulting fees from Gilead, Merck Sharp & Dohme, and ViiV Healthcare, all outside the submitted work. C. C. has received honoraria and travel grants from Gilead, ViiV Health Care, and MSD (Merck) and personal consulting fees from ViiV Health Care all outside the submitted work. All other authors report no potential conflicts.
Figures
References
-
- Landman R, de Truchis P, Assoumou L, et al. A 4-days-on and 3-days-off maintenance treatment strategy for adults with HIV-1 (ANRS 170 QUATUOR): a randomised, open-label, multicentre, parallel, non-inferiority trial. Lancet HIV 2022; 9:e79–90. - PubMed
-
- Haute Autorité de Santé. VOCABRIA (cabotégravir)—VIH. Available at: https://www.has-sante.fr/jcms/p_3412898/fr/vocabria-cabotegravir-vih. Accessed 10 March 2024.
-
- Orkin C, Schapiro JM, Perno CF, et al. Expanded multivariable models to assist patient selection for long-acting cabotegravir + rilpivirine treatment: clinical utility of a combination of patient, drug concentration, and viral factors associated with virologic failure. Clin Infect Dis 2023; 7:1423–31. - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

