Glucose-6-phosphate dehydrogenase regulates mitophagy by maintaining PINK1 stability
- PMID: 39872984
- PMCID: PMC11749863
- DOI: 10.1093/lifemeta/loae040
Glucose-6-phosphate dehydrogenase regulates mitophagy by maintaining PINK1 stability
Abstract
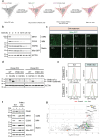
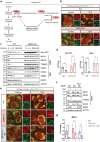
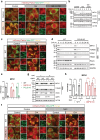
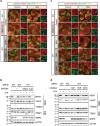
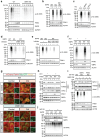
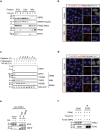
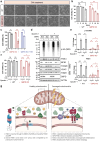
Glucose-6-phosphate dehydrogenase (G6PD) is the rate-limiting enzyme in the pentose phosphate pathway (PPP) in glycolysis. Glucose metabolism is closely implicated in the regulation of mitophagy, a selective form of autophagy for the degradation of damaged mitochondria. The PPP and its key enzymes such as G6PD possess important metabolic functions, including biosynthesis and maintenance of intracellular redox balance, while their implication in mitophagy is largely unknown. Here, via a whole-genome CRISPR-Cas9 screening, we identified that G6PD regulates PINK1 (phosphatase and tensin homolog [PTEN]-induced kinase 1)-Parkin-mediated mitophagy. The function of G6PD in mitophagy was verified via multiple approaches. G6PD deletion significantly inhibited mitophagy, which can be rescued by G6PD reconstitution. Intriguingly, while the catalytic activity of G6PD is required, the known PPP functions per se are not involved in mitophagy regulation. Importantly, we found a portion of G6PD localized at mitochondria where it interacts with PINK1. G6PD deletion resulted in an impairment in PINK1 stabilization and subsequent inhibition of ubiquitin phosphorylation, a key starting point of mitophagy. Finally, we found that G6PD deletion resulted in lower cell viability upon mitochondrial depolarization, indicating the physiological function of G6PD-mediated mitophagy in response to mitochondrial stress. In summary, our study reveals a novel role of G6PD as a key positive regulator in mitophagy, which bridges several important cellular processes, namely glucose metabolism, redox homeostasis, and mitochondrial quality control.
Keywords: G6PD; NADPH; PINK1; PPP; ROS; mitophagy.
© The Author(s) 2024. Published by Oxford University Press on behalf of Higher Education Press.
Conflict of interest statement
H.M.S. holds the position of Editorial Board Member for Life Metabolism and is blinded from reviewing or making decisions for the manuscript. The other authors declare that no conflict of interest exists.
Figures
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
