Glia Modulates Immune Responses in the Retina Through Distinct MHC Pathways
- PMID: 39873321
- PMCID: PMC11845847
- DOI: 10.1002/glia.24656
Glia Modulates Immune Responses in the Retina Through Distinct MHC Pathways
Abstract
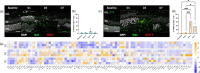
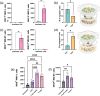
Glia antigen-presenting cells (APCs) are pivotal regulators of immune surveillance within the retina, maintaining tissue homeostasis and promptly responding to insults. However, the intricate mechanisms underlying their local coordination and activation remain unclear. Our study integrates an animal model of retinal injury, retrospective analysis of human retinas, and in vitro experiments to gain insights into the crucial role of antigen presentation in neuroimmunology during retinal degeneration (RD), uncovering the involvement of various glial cells, notably Müller glia and microglia. Glial cells act as sentinels, detecting antigens released during degeneration and interacting with T-cells via MHC molecules, which are essential for immune responses. Microglia function as APCs via the MHC Class II pathway, upregulating key molecules such as Csf1r and cytokines. In contrast, Müller cells act through the MHC Class I pathway, exhibiting upregulated antigen processing genes and promoting a CD8+ T-cell response. Distinct cytokine signaling pathways, including TNF-α and IFN Type I, contribute to the immune balance. Human retinal specimens corroborate these findings, demonstrating glial activation and MHC expression correlating with degenerative changes. In vitro assays also confirmed differential T-cell migration responses to activated microglia and Müller cells, highlighting their role in shaping the immune milieu within the retina. In summary, our study emphasizes the involvement of retinal glial cells in modulating the immune response after insults to the retinal parenchyma. Unraveling the intricacies of glia-mediated antigen presentation in RD is essential for developing precise therapeutic interventions for retinal pathologies.
Keywords: Müller cells; T‐cells; antigen‐presenting cells; microglia; retina.
© 2025 The Author(s). GLIA published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Prohibitin 2 deficiency in photoreceptors leads to progressive retinal degeneration and facilitated Müller glia engulfing microglia debris.Exp Eye Res. 2024 Jul;244:109935. doi: 10.1016/j.exer.2024.109935. Epub 2024 May 17. Exp Eye Res. 2024. PMID: 38763352
-
Patterns of NFkB activation resulting from damage, reactive microglia, cytokines, and growth factors in the mouse retina.Exp Neurol. 2023 Jan;359:114233. doi: 10.1016/j.expneurol.2022.114233. Epub 2022 Sep 27. Exp Neurol. 2023. PMID: 36174748 Free PMC article.
-
Retinal microglia signaling affects Müller cell behavior in the zebrafish following laser injury induction.Glia. 2019 Jun;67(6):1150-1166. doi: 10.1002/glia.23601. Epub 2019 Feb 22. Glia. 2019. PMID: 30794326
-
Microglia activation in retinal degeneration.J Leukoc Biol. 2007 Jun;81(6):1345-51. doi: 10.1189/jlb.0207114. Epub 2007 Apr 3. J Leukoc Biol. 2007. PMID: 17405851 Review.
-
Müller glial cell reprogramming and retina regeneration.Nat Rev Neurosci. 2014 Jul;15(7):431-42. doi: 10.1038/nrn3723. Epub 2014 Jun 4. Nat Rev Neurosci. 2014. PMID: 24894585 Free PMC article. Review.
Cited by
-
Immune cell contribution to vascular complications in diabetes.Front Endocrinol (Lausanne). 2025 May 21;16:1549945. doi: 10.3389/fendo.2025.1549945. eCollection 2025. Front Endocrinol (Lausanne). 2025. PMID: 40469434 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

