Frequently arising ESX-1-associated phase variants influence Mycobacterium tuberculosis fitness in the presence of host and antibiotic pressures
- PMID: 39873486
- PMCID: PMC11898584
- DOI: 10.1128/mbio.03762-24
Frequently arising ESX-1-associated phase variants influence Mycobacterium tuberculosis fitness in the presence of host and antibiotic pressures
Abstract
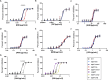
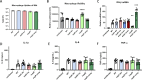
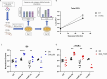
Mycobacterium tuberculosis (Mtb) exhibits an impressive ability to adapt to rapidly changing environments, despite its genome's apparent stability. Recently, phase variation through indel formation in homopolymeric tracts (HT) has emerged as a potentially important mechanism promoting adaptation in Mtb. This study examines the impact of common phase variants associated with the ESX-1 type VII secretion system, focusing on a highly variable HT upstream of the ESX-1 regulatory factor, espR. By engineering this frequently observed indel into an isogenic background, we demonstrate that a single nucleotide insertion in the espR 5'UTR causes post-transcriptional upregulation of EspR protein abundance and corresponding alterations in the EspR regulon. Consequently, this mutation increases the expression of ESX-1 components in the espACD operon and enhances ESX-1 substrate secretion. We find that this indel specifically increases isoniazid resistance without impacting the effectiveness of other drugs tested. Furthermore, we show that two distinct observed HT indels that regulate either espR translation or espACD transcription increase bacterial fitness in a mouse infection model. The presence of multiple ESX-1-associated HTs provides a mechanism to combinatorially tune protein secretion, drug sensitivity, and host-pathogen interactions. More broadly, these findings support emerging data that Mtb utilizes HT-mediated phase variation to direct genetic variation to certain sites across the genome in order to adapt to changing pressures.
Importance: Mycobacterium tuberculosis (Mtb) is responsible for more deaths worldwide than any other single infectious agent. Understanding how this pathogen adapts to the varied environmental pressures imposed by host immunity and antibiotics has important implications for the design of more effective therapies. In this work, we show that the genome of Mtb contains multiple contingency loci that control the activity of the ESX-1 secretion system, which is critical for interactions with the host. These loci consist of homopolymeric DNA tracts in gene regulatory regions that are subject to high-frequency reversible variation and act to tune the activity of ESX-1. We find that variation at these sites increases the fitness of Mtb in the presence of antibiotic and/or during infection. These findings indicate that Mtb has the ability to diversify its genome in specific sites to create subpopulations of cells that are preadapted to new conditions.
Keywords: bacterial genetics; phase variation; tuberculosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
- F32AI174653/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- P01 AI143575/AI/NIAID NIH HHS/United States
- F32 AI174653/AI/NIAID NIH HHS/United States
- AI181898/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- AI162598/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- P01 AI181898/AI/NIAID NIH HHS/United States
- AI156415/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- AI143575/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- U19 AI162598/AI/NIAID NIH HHS/United States
- R21 AI156415/AI/NIAID NIH HHS/United States
- T32A100734/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources

