Genetic diversity and virulence of Bacillus cereus group isolates from bloodstream infections
- PMID: 39873504
- PMCID: PMC11878096
- DOI: 10.1128/spectrum.02407-24
Genetic diversity and virulence of Bacillus cereus group isolates from bloodstream infections
Erratum in
-
Erratum for Okutani et al., "Genetic diversity and virulence of Bacillus cereus group isolates from bloodstream infections".Microbiol Spectr. 2025 Nov 4;13(11):e0293725. doi: 10.1128/spectrum.02937-25. Epub 2025 Oct 10. Microbiol Spectr. 2025. PMID: 41071094 Free PMC article. No abstract available.
Abstract
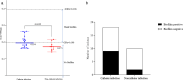
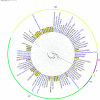
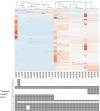
Bacillus cereus catheter-related bloodstream infections (CRBSIs) are an increasing concern in Japanese hospitals. Although their clinical characteristics have been explored, the genetic relationships and virulence profiles of B. cereus isolates from CRBSIs remain understudied. Here, using advanced genomic techniques, we investigated the genetic diversity, phylogenetic relationships, and virulence profiles of B. cereus isolates from patients with bloodstream infections. We analyzed 28 B. cereus group strains isolated from blood samples at the University of Tokyo Hospital between 2005 and 2017 using whole-genome sequencing, core-genome single-nucleotide polymorphism (SNP) typing, and virulence gene profiling. Core-genome SNP analysis revealed significant genetic diversity among the isolates, suggesting multiple independent sources of infection. The isolates predominantly belonged to panC clades III and IV, with distinct virulence gene profiles. All panC clade III isolates contained hbl operon genes, whereas four isolates from clade IV harbored cereulide synthetase genes (cesABCD). One isolate possessed a capsule gene operon (capBCADE), a rare finding among clinical B. cereus strains. Biofilm formation ability was observed in 50% of catheter-related isolates, although this ability was not significantly different from that of the noncatheter-related isolates.IMPORTANCEThis study provides novel insights into the genetic diversity and virulence potential of B. cereus strains causing bloodstream infections in a Japanese hospital setting. These findings suggest diverse infection pathways and highlight the importance of continuous molecular epidemiological surveillance for effective infection control.
Keywords: Bacillus cereus group; bloodstream infection; epidemiology; whole-genome analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Okamoto A, Okutani A. 2024. The Bacillus cereus group, p 957–986. In Yi-Wei Tang MH, Dongyou L, Sails A, Spearman P, Zhang JR (ed), Molecular medical microbiology. Elsevier, London.
-
- Glasset B, Herbin S, Granier SA, Cavalié L, Lafeuille E, Guérin C, Ruimy R, Casagrande-Magne F, Levast M, Chautemps N, Decousser J-W, Belotti L, Pelloux I, Robert J, Brisabois A, Ramarao N. 2018. Bacillus cereus, a serious cause of nosocomial infections: epidemiologic and genetic survey. PLoS One 13:e0194346. doi: 10.1371/journal.pone.0194346 - DOI - PMC - PubMed
-
- Mills E, Sullivan E, Kovac J. 2022. Comparative analysis of Bacillus cereus group isolates’ resistance using disk diffusion and broth microdilution and the correlation between antimicrobial resistance phenotypes and genotypes. Appl Environ Microbiol 88:e0230221. doi: 10.1128/aem.02302-21 - DOI - PMC - PubMed
-
- Aoyagi T, Oshima K, Endo S, Baba H, Kanamori H, Yoshida M, Tokuda K, Kaku M. 2020. Ba813 harboring Bacillus cereus, genetically closely related to Bacillus anthracis, causing nosocomial bloodstream infection: bacterial virulence factors and clinical outcome. PLoS One 15:e0235771. doi: 10.1371/journal.pone.0235771 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

