Systematic analyses uncover robust salivary microbial signatures and host-microbiome perturbations in oral squamous cell carcinoma
- PMID: 39873508
- PMCID: PMC11834404
- DOI: 10.1128/msystems.01247-24
Systematic analyses uncover robust salivary microbial signatures and host-microbiome perturbations in oral squamous cell carcinoma
Abstract
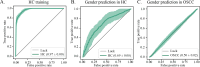
Oral squamous cell carcinoma (OSCC) is a prevalent malignancy in the oral-maxillofacial region with a poor prognosis. Oral microbiomes play a potential role in the pathogenesis of this disease. However, findings from individual studies have been inconsistent, and a comprehensive understanding of OSCC-associated microbiome dysbiosis remains elusive. Here, we conducted a large-scale meta-analysis by integrating 11 publicly available data sets comprising salivary microbiome profiles of OSCC patients and healthy controls. After correcting for batch effects, we observed significantly elevated alpha diversity and distinct beta-diversity patterns in the OSCC salivary microbiome compared to healthy controls. Leveraging random effects models, we identified robust microbial signatures associated with OSCC across data sets, including enrichment of taxa such as Streptococcus, Lactobacillus, Prevotella, Bulleidia moorei, and Haemophilus in OSCC samples. The machine learning models constructed from these microbial markers accurately predicted OSCC status, highlighting their potential as non-invasive diagnostic biomarkers. Intriguingly, our analyses revealed that the age- and gender-associated signatures in normal saliva microbiome were disrupted in the OSCC, suggesting perturbations in the intricate host-microbe interactions. Collectively, our findings uncovered complex alterations in the oral microbiome in OSCC, providing novel insights into disease etiology and paving the way for microbiome-based diagnostic and therapeutic strategies. Given that the salivary microbiome can reflect the overall health status of the host and that saliva sampling is a safe, non-invasive approach, it may be worthwhile to conduct broader screening of the salivary microbiome in high-risk OSCC populations as implications for early detection.
Importance: The oral cavity hosts a diverse microbial community that plays a crucial role in systemic and oral health. Accumulated research has investigated significant differences in the saliva microbiota associated with oral cancer, suggesting that microbiome dysbiosis may contribute to the pathogenesis of oral squamous cell carcinoma (OSCC). However, the specific microbial alterations linked to OSCC remain controversial. This meta-analysis reveals robust salivary microbiome alterations. Machine learning models using differential operational taxonomic units accurately predicted OSCC status, highlighting the potential of the salivary microbiome as a non-invasive diagnostic biomarker. Interestingly, age- and gender-associated signatures in the normal salivary microbiome were disrupted in OSCC, suggesting perturbations in host-microbe interactions.
Keywords: age; gender; meta-analysis; oral squamous cell carcinoma; salivary microbiome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The oral microbiome and its role in oral squamous cell carcinoma: a systematic review of microbial alterations and potential biomarkers.Pathologica. 2024 Dec;116(6):338-357. doi: 10.32074/1591-951X-N867. Pathologica. 2024. PMID: 39748720
-
Bioinformatics identification and validation of m6A/m1A/m5C/m7G/ac4 C-modified genes in oral squamous cell carcinoma.BMC Cancer. 2025 Jul 1;25(1):1055. doi: 10.1186/s12885-025-14216-7. BMC Cancer. 2025. PMID: 40597017 Free PMC article.
-
Oral Microbial Dysbiosis Driven by Periodontitis Facilitates Oral Squamous Cell Carcinoma Progression.Cancers (Basel). 2025 Jun 28;17(13):2181. doi: 10.3390/cancers17132181. Cancers (Basel). 2025. PMID: 40647479 Free PMC article.
-
Systematic review on oral microbial dysbiosis and its clinical associations with head and neck squamous cell carcinoma.Head Neck. 2023 Aug;45(8):2120-2135. doi: 10.1002/hed.27422. Epub 2023 May 30. Head Neck. 2023. PMID: 37249085
-
Bioinformatics-driven salivary microbial and functional profiling for identifying biomarkers in oral cancer and tobacco abusers in the Indian population.Arch Oral Biol. 2025 Jun 30;178:106346. doi: 10.1016/j.archoralbio.2025.106346. Online ahead of print. Arch Oral Biol. 2025. PMID: 40616950
Cited by
-
Oral and intratumoral microbiota influence tumor immunity and patient survival.Front Immunol. 2025 May 21;16:1572152. doi: 10.3389/fimmu.2025.1572152. eCollection 2025. Front Immunol. 2025. PMID: 40475759 Free PMC article.
-
Gastric Cancer and Microbiota: Exploring the Microbiome's Role in Carcinogenesis and Treatment Strategies.Life (Basel). 2025 Jun 23;15(7):999. doi: 10.3390/life15070999. Life (Basel). 2025. PMID: 40724502 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
