The UCP2/PINK1/LC3b-mediated mitophagy is involved in the protection of NRG1 against myocardial ischemia/reperfusion injury
- PMID: 39874927
- PMCID: PMC11808529
- DOI: 10.1016/j.redox.2025.103511
The UCP2/PINK1/LC3b-mediated mitophagy is involved in the protection of NRG1 against myocardial ischemia/reperfusion injury
Abstract
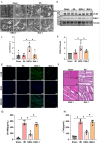
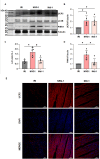
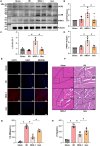
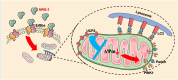
Available evidence indicates that neuregulin-1 (NRG-1) can provide a protection against myocardial ischemia/reperfusion (I/R) injury and is involved in various cardioprotective interventions by potential regulation of mitophagy. However, the molecular mechanisms linking NRG-1 and mitophagy remain to be clarified. In this study, both an in vivo myocardial I/R injury model of rats and an in vitro hypoxia/reoxygenation (H/R) model of H9C2 cardiomyocytes were applied to determine whether NRG-1 postconditioning attenuated myocardial I/R injury through the regulation of mitophagy and to explore the underlying mechanisms. In the in vivo experiment, cardioprotective effects of NRG-1 were determined by infarct size, cardiac enzyme and histopathologic examinations. The potential downstream signaling pathways and molecular targets of NRG-1 were screened by the RNA sequencing and the Protein-Protein Interaction Networks. The expression levels of mitochondrial uncoupling protein 2 (UCP2) and mitophagy-related proteins in both the I/R myocardium and H/R cardiomyocytes were measured by immunofluorescence staining and Western blots. The activation of mitophagy was observed with transmission electron microscopy and JC-1 staining. The KEGG and GSEA analyses showed that the mitophagy-related signaling pathways were enriched in the I/R myocardium treated with NRG-1, and UCP2 exhibited a significant correlation between mitophagy and interaction with PINK1. Meanwhile, the treatment with mitophagy inhibitor Mdivi-1 significant eliminated the cardioprotective effects of NRG-1 postconditioning in vivo, and the challenge with UCP2 inhibitor genipin could also attenuate the activating effect of NRG-1 postconditioning on mitophagy. Consistently, the in vitro experiment using H9C2 cardiomyocytes showd that NRG-1 treatment significantly up-regulated the expression levels of UCP2 and mitophagy-related proteins, and activated the mitophagy, whereas the challenge with small interfering RNA-mediated UCP2 knockdown abolished the effects of NRG-1. Thus, it is conclused that NRG-1 postconditioning can produce a protection against the myocardial I/R injury by activating mitophagy through the UCP2/PINK1/LC3B signaling pathway.
Keywords: Ischemia/reperfusion injury; Mitophagy; Neuregulin-1; Uncoupling protein 2.
Copyright © 2025 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors have declared no conflict of interest.
Figures





References
-
- Galli M., Niccoli G., De Maria G., Brugaletta S., Montone R.A., Vergallo R., Benenati S., Magnani G., D'Amario D., Porto I., et al. Coronary microvascular obstruction and dysfunction in patients with acute myocardial infarction. Nat. Rev. Cardiol. 2024;21:283–298. doi: 10.1038/s41569-023-00953-4. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

