Activation of lysophagy by a TBK1-SCFFBXO3-TMEM192-TAX1BP1 axis in response to lysosomal damage
- PMID: 39875384
- PMCID: PMC11775327
- DOI: 10.1038/s41467-025-56294-y
Activation of lysophagy by a TBK1-SCFFBXO3-TMEM192-TAX1BP1 axis in response to lysosomal damage
Abstract
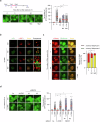
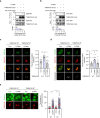
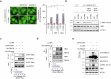
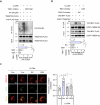
Lysophagy eliminates damaged lysosomes and is crucial to cellular homeostasis; however, its underlying mechanisms are not entirely understood. We screen a ubiquitination-related compound library and determine that the substrate recognition component of the SCF-type E3 ubiquitin ligase complex, SCFFBXO3(FBXO3), which is a critical lysophagy regulator. Inhibition of FBXO3 reduces lysophagy and lysophagic flux in response to L-leucyl-L-leucine methyl ester (LLOMe). Furthermore, FBXO3 interacts with TMEM192, leading to its ubiquitination in LLOMe-treated cells. We also identify TAX1BP1 as a critical autophagic adaptor that recognizes ubiquitinated TMEM192 during lysophagy and find that TBK1 activation is crucial for lysophagy, as it phosphorylates FBXO3 in response to lysosomal damage. Knockout of FBXO3 significantly impairs lysophagy, and its reconstitution with a loss-of-function mutant (V221I) further confirms its essential role in lysophagy regulation. Collectively, our findings highlight the significance of the TBK1-FBXO3-TMEM192-TAX1BP1 axis in lysophagy and emphasize the critical role of FBXO3 in lysosomal integrity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Saftig, P. & Klumperman, J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol.10, 623–635 (2009). - PubMed
MeSH terms
Substances
Grants and funding
- P0025489/Korea Institute for Advancement of Technology (KIAT)
- RS-2024-00338475/National Research Foundation of Korea (NRF)
- RS-2024-00453488/National Research Foundation of Korea (NRF)
- RS-2024-00463344/National Research Foundation of Korea (NRF)
- RS-2023-00301914/National Research Foundation of Korea (NRF)
LinkOut - more resources
Full Text Sources
Miscellaneous

