Associations between common genetic variants and income provide insights about the socio-economic health gradient
- PMID: 39875632
- PMCID: PMC12018258
- DOI: 10.1038/s41562-024-02080-7
Associations between common genetic variants and income provide insights about the socio-economic health gradient
Abstract
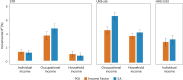
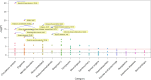
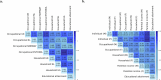
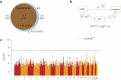
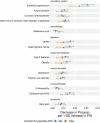
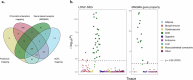
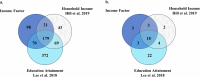
We conducted a genome-wide association study on income among individuals of European descent (N = 668,288) to investigate the relationship between socio-economic status and health disparities. We identified 162 genomic loci associated with a common genetic factor underlying various income measures, all with small effect sizes (the Income Factor). Our polygenic index captures 1-5% of income variance, with only one fourth due to direct genetic effects. A phenome-wide association study using this index showed reduced risks for diseases including hypertension, obesity, type 2 diabetes, depression, asthma and back pain. The Income Factor had a substantial genetic correlation (0.92, s.e. = 0.006) with educational attainment. Accounting for the genetic overlap of educational attainment with income revealed that the remaining genetic signal was linked to better mental health but reduced physical health and increased risky behaviours such as drinking and smoking. These findings highlight the complex genetic influences on income and health.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Ridley, M., Rao, G., Schilbach, F. & Patel, V. Poverty, depression, and anxiety: causal evidence and mechanisms. Science370, eaay0214 (2020). - PubMed
-
- Stevenson, B. & Wolfers, J. Subjective well-being and income: is there any evidence of satiation? Am. Econ. Rev.103, 598–604 (2013).
-
- Wilkinson, R. G. & Marmot, M. Social Determinants of Health: The Solid Facts (World Health Organization, 2003).
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

