Molecular Profiling of Odontoclasts during Physiological Tooth Replacement
- PMID: 39876039
- PMCID: PMC12000629
- DOI: 10.1177/00220345241304756
Molecular Profiling of Odontoclasts during Physiological Tooth Replacement
Erratum in
-
Corrigendum to Molecular Profiling of Odontoclasts during Physiological Tooth Replacement.J Dent Res. 2025 May;104(5):572-573. doi: 10.1177/00220345251322122. Epub 2025 Feb 13. J Dent Res. 2025. PMID: 39945328 Free PMC article. No abstract available.
-
Corrigendum to Molecular Profiling of Odontoclasts during Physiological Tooth Replacement.J Dent Res. 2025 Dec;104(13):1567. doi: 10.1177/00220345251384972. Epub 2025 Oct 3. J Dent Res. 2025. PMID: 41042130 Free PMC article. No abstract available.
Abstract
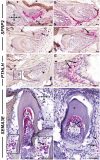
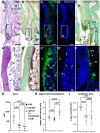
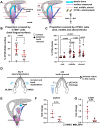
The odontoclast is a rarely studied cell type that is overly active in many dental pathologies, leading to tooth loss. It is difficult to find diphyodont mammals in which either physiological or pathological root resorption can be studied. Here we use the adult leopard gecko, which has repeated cycles of physiological tooth resorption and shedding. RNA-seq was carried out to compare gene expression profiles of functional teeth to developing teeth. Genes more highly expressed in bell-stage developing teeth were related to morphogenesis (PTHLH, SFRP2, SHH, EDAR). Some genes expressed in osteoclasts (ACP5, CTSK, CSF1R) were relatively more abundant in functional teeth compared with developing teeth. There was, however, no differential expression of RANKL (TNFSF11) in the 2 tooth types. In addition, functional teeth expressed proteolysis genes not found in osteoclasts (ADAMTS2, 3, 4, 14; CTSA, CTSH, CTSS). We used tartrate acid resistant phosphatase and cathepsin K (CTSK) staining to identify odontoclasts in and around the gecko dentition. There were 3 populations of CTSK cells: (1) large, functional multinucleated odontoclasts in the crown of the tooth with a ruffled border inside resorption pits; (2) smaller, precursor cells in the pulp with fewer nuclei; and (3) flattened external precursor cells next to the root and bone of attachment. We found a positive relationship between developing teeth and the population of CTSK+ cells on the root surface. We tested a candidate signal that may be involved in CTSK+ cell presence. An antagonist of CSF1R was delivered to developing teeth in vivo, which resulted in a significant decrease in CTSK and CSF1R compared with DMSO controls. Thus, the CSF1 signaling pathway is upstream of CTSK in teeth. This is the first work to detail the molecular characteristics of odontoclasts during physiological tooth shedding and to demonstrate that in vivo, local drug delivery is possible in the gecko model.
Keywords: RNA-seq; cathepsin K; gecko; polyphyodonty; reptile; root resorption.
Conflict of interest statement
Declaration of Conflicting InterestsThe authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures

References
-
- Alyahya L, Myers GL. 2021. Denosumab use as a predictor variable for external cervical resorption: a case-control study. J Endod. 47(3):366–373. - PubMed
-
- Bjerklin K, Al-Najjar M, Karestedt H, Andren A. 2008. Agenesis of mandibular second premolars with retained primary molars: a longitudinal radiographic study of 99 subjects from 12 years of age to adulthood. Eur J Orthod. 30(3):254–261. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

