Meta-analysis of retinal transcriptome profiling studies in animal models of myopia
- PMID: 39876870
- PMCID: PMC11772478
- DOI: 10.3389/fmed.2024.1479891
Meta-analysis of retinal transcriptome profiling studies in animal models of myopia
Abstract
Objective: Myopia prevalence is increasing at alarming rates, yet the underlying mechanistic causes are not understood. Several studies have employed experimental animal models of myopia and transcriptome profiling to identify genes and pathways contributing to myopia. In this study, we determined the retinal transcriptome changes in response to form deprivation in mouse retinas. We then conducted a transcriptome meta-analysis incorporating all publicly available datasets and analyzed how the results related to the genes associated with refractive errors in human genome-wide association studies (GWAS).
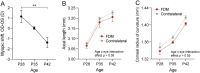
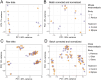
Methods: Form deprivation was induced in three male C57BL6/J mice from postnatal day 28 (P28) to P42. Retinal gene expression was analyzed with RNA sequencing, followed by differential gene expression analysis with DESeq2 and identification of associated pathways with the Kyoto Encyclopedia of Genes and Genomes (KEGG). A systematic search identified four similar retinal transcriptomics datasets in response to experimental myopia using chicks or mice. The five studies underwent transcriptome meta-analyses to determine retinal gene expression changes and associated pathways. The results were compared with genes associated with human myopia.
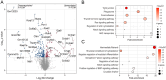
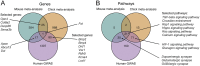
Results: Differential gene expression analysis of form-deprived mouse retinas revealed 235 significantly altered transcripts, implicating the BMP2 signaling pathway and circadian rhythms, among others. Transcriptome-wide meta-analyses of experimental myopia datasets found 427 differentially expressed genes in the mouse model and 1,110 in the chick model, with limited gene overlap between species. Pathway analysis of these two gene sets implicated TGF-beta signaling and circadian rhythm pathways in both mouse and chick retinas. Some pathways associated only with mouse retinal changes included dopamine signaling and HIF-1 signaling pathway, whereas glucagon signaling was only associated with gene changes in chick retinas. The follistatin gene changed in both mouse and chick retinas and has also been implicated in human myopia. TGF-beta signaling pathway and circadian entrainment processes were associated with myopia in mice, chicks, and humans.
Conclusion: This study highlights the power of combining datasets to enhance statistical power and identify robust gene expression changes across different experimental animal models and conditions. The data supports other experimental evidence that TGF-beta signaling pathway and circadian rhythms are involved in myopic eye growth.
Keywords: RNA-Seq; experimental models; meta-analysis; myopia; transcriptomics.
Copyright © 2025 Palumaa, Balamurugan and Pardue.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources

