A chromosome-level, haplotype-resolved genome assembly and annotation for the Eurasian minnow (Leuciscidae: Phoxinus phoxinus) provide evidence of haplotype diversity
- PMID: 39877992
- PMCID: PMC11775470
- DOI: 10.1093/gigascience/giae116
A chromosome-level, haplotype-resolved genome assembly and annotation for the Eurasian minnow (Leuciscidae: Phoxinus phoxinus) provide evidence of haplotype diversity
Abstract
Background: In this study, we present an in-depth analysis of the Eurasian minnow (Phoxinus phoxinus) genome, highlighting its genetic diversity, structural variations, and evolutionary adaptations. We generated an annotated haplotype-phased, chromosome-level genome assembly (2n = 50) by integrating high-fidelity (HiFi) long reads and chromosome conformation capture data (Hi-C).
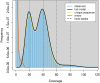
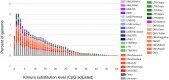
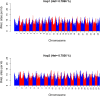
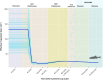
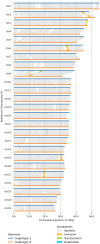
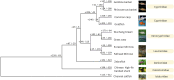
Results: We achieved a haploid size of 940 megabase pairs (Mbp) for haplome 1 and 929 Mbp for haplome 2 with high scaffold N50 values of 36.4 Mb and 36.6 Mb and BUSCO scores of 96.9% and 97.2%, respectively, indicating a highly complete genome assembly. We detected notable heterozygosity (1.43%) and a high repeat content (approximately 54%), primarily consisting of DNA transposons, which contribute to genome rearrangements and variations. We found substantial structural variations within the genome, including insertions, deletions, inversions, and translocations. These variations affect genes enriched in functions such as dephosphorylation, developmental pigmentation, phagocytosis, immunity, and stress response. In the annotation of protein-coding genes, 30,980 messenger RNAs and 23,497 protein-coding genes were identified with a high completeness score, which further underpins the high contiguity of our genome assemblies. We performed a gene family evolution analysis by comparing our proteome to 10 other teleost species, which identified immune system gene families that prioritize histone-based disease prevention over NB-LRR-related-based immune responses. Additionally, demographic analysis indicates historical fluctuations in the effective population size of P. phoxinus, likely correlating with past climatic changes.
Conclusions: This annotated, phased reference genome provides a crucial resource for resolving the taxonomic complexity within the genus Phoxinus and highlights the importance of haplotype-phased assemblies in understanding haplotype diversity in species characterized by high heterozygosity.
Keywords: Phoxinus; annotation; comparative genomics; haplotype-phased genome assembly; immune system; structural variants; transposable elements.
© The Author(s) 2025. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Kottelat M, Freyhof J. Handbook of European freshwater fishes. Ichthyol Res. 2008;55:99. 10.1007/s10228-007-0012-3. - DOI
-
- Kottelat M. Three new species of Phoxinus from Greece and southern France (Teleostei: cyprinidae). Ichthyol Explor Freshw. 2007;18(2):145–62.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

