Epigenetic state and gene expression remain stable after CRISPR/Cas-mediated chromosomal inversions
- PMID: 39878102
- PMCID: PMC11840415
- DOI: 10.1111/nph.20403
Epigenetic state and gene expression remain stable after CRISPR/Cas-mediated chromosomal inversions
Abstract
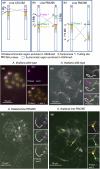
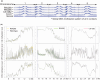
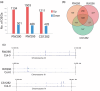
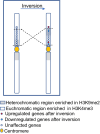
The epigenetic state of chromatin, gene activity and chromosomal positions are interrelated in plants. In Arabidopsis thaliana, chromosome arms are DNA-hypomethylated and enriched with the euchromatin-specific histone mark H3K4me3, while pericentromeric regions are DNA-hypermethylated and enriched with the heterochromatin-specific mark H3K9me2. We aimed to investigate how the chromosomal location affects epigenetic stability and gene expression by chromosome engineering. Two chromosomal inversions of different sizes were induced using CRISPR/Cas9 to move heterochromatic, pericentric sequences into euchromatic regions. The epigenetic status of these lines was investigated using whole-genome bisulfite sequencing and chromatin immunoprecipitation. Gene expression changes following the induction of the chromosomal inversions were studied via transcriptome analysis. Both inversions had a minimal impact on the global distribution of histone marks and DNA methylation patterns, although minor epigenetic changes were observed across the genome. Notably, the inverted chromosomal regions and their borders retained their original epigenetic profiles. Gene expression analysis showed that only 0.5-1% of genes were differentially expressed genome-wide following the induction of the inversions. CRISPR/Cas-induced chromosomal inversions minimally affect epigenetic landscape and gene expression, preserving their profiles in subsequent generations.
Keywords: CRISPR/Cas; chromosome engineering; epigenetics; gene expression; inversion.
© 2025 The Author(s). New Phytologist © 2025 New Phytologist Foundation.
Conflict of interest statement
None declared.
Figures

Similar articles
-
Efficient induction of heritable inversions in plant genomes using the CRISPR/Cas system.Plant J. 2019 May;98(4):577-589. doi: 10.1111/tpj.14322. Epub 2019 Apr 16. Plant J. 2019. PMID: 30900787
-
Changing local recombination patterns in Arabidopsis by CRISPR/Cas mediated chromosome engineering.Nat Commun. 2020 Sep 4;11(1):4418. doi: 10.1038/s41467-020-18277-z. Nat Commun. 2020. PMID: 32887885 Free PMC article.
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
-
Molecular landscape of modified histones in Drosophila heterochromatic genes and euchromatin-heterochromatin transition zones.PLoS Genet. 2008 Jan;4(1):e16. doi: 10.1371/journal.pgen.0040016. Epub 2007 Dec 13. PLoS Genet. 2008. PMID: 18208336 Free PMC article.
-
dCas-Based Tools to Visualize Chromatin or Modify Epigenetic Marks at Specific Plant Genomic Loci.Methods Mol Biol. 2025;2873:305-332. doi: 10.1007/978-1-0716-4228-3_17. Methods Mol Biol. 2025. PMID: 39576609 Review.
Cited by
-
Thriving or Withering? Plant Molecular Cytogenetics in the First Quarter of the 21st Century.Int J Mol Sci. 2025 Jul 21;26(14):7013. doi: 10.3390/ijms26147013. Int J Mol Sci. 2025. PMID: 40725259 Free PMC article. Review.
-
Chromosome engineering points to the cis-acting mechanism of chromosome arm-specific telomere length setting and robustness of plant phenotype, chromatin structure and gene expression.Plant J. 2025 Feb;121(4):e70024. doi: 10.1111/tpj.70024. Plant J. 2025. PMID: 39962352 Free PMC article.
References
-
- Beying N, Schmidt C, Pacher M, Houben A, Puchta H. 2020. CRISPR–Cas9‐mediated induction of heritable chromosomal translocations in Arabidopsis. Nature Plants 6: 638–645. - PubMed
-
- Boideau F, Richard G, Coriton O, Huteau V, Belser C, Deniot G, Eber F, Falentin C, Ferreira de Carvalho J, Gilet M et al. 2022. Epigenomic and structural events preclude recombination in Brassica napus . New Phytologist 234: 545–559. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

